THE GIFA PROGRAM.VERSION 4.0
This program has been developed in several NMR groups in France, first in I.C.S.N. in Gif/Yvette, then in Ecole Polytechnique in Palaiseau and now in Montpellier under the coordination of Marc-André Delsuc. Please contact :
M.A.Delsuc
Centre de Biochimie Structurale
Faculté de Pharmacie,
15 avenue Charles Flahault
34060 Montpellier Cedex FRANCE.
E-Mail : mad@cbs.univ-montp1.fr (Internet)
Tel : (33) 67 04 34 36 (In case of emergency)
The program includes all the classical processing, displaying and plotting
capabilities of an NMR program, as well as more advanced processing techniques
such as Maximum Entropy processing or Linear prediction. There is no actual
limit to the size of the data-set which can be processed. A complete macro
language permits to builds sophisticated processing. The Graphic User Interface
is fully designable and programmable by the user.
The program currently runs on several UNIX platforms. The graphics can be
displayed on X-Windows terminals. Plots are generated on postscript or HP-GL
devices.
This version V4.0 is now in final version, however it is evoluating all the
time, check your ftp server ! This version is distributed under a licence.
There is no fees for academic laboratories, however the licence requires that
you refer to Gifa in any published work which depend in some manner on the Gifa
program.
This program can be downloaded by anonymous FTP on Internet, the home site is
the following address : ftp://tome.cbs.univ-montp1.fr/pub/gifa_v4 .
Previous released versions have been : 1.0 (Initial VMS Version), 1.1 (revised
version), 2.0 (first port to UNIX), 2.1 (first stabilised version), 2.5
(introduction of linear prediction, polynomial baseline correction), 3.0
(introduction of macro-language, on-file processing, etc...), 3.1 (new
peak-picker and line-fitter), 4.0 (Motif version, new user interface, new
language capabilities, new cache, extended line fitter). Several versions have
been used internally in our lab, and may have been released by mistake.
*Full array of apodisation functions (shifted sine-bells and squared
sine-bells; exponential; Gaussian; trapezoid, and combinations).
*Fast Fourier transform (direct and inverse; real, complex, and hypercomplex).
Hilbert transforms.
*Zero-filling and truncation.
*Phasing of 1D, 2D and 3D spectra.
*Processing in memory (for regular data size) or on disk (for very large
data-set) with similar syntaxes
*Non-limited size data-set with optimised file access protocol.
*Very fast processing :
* 2'30" on Macintosh Centris 650 - running MachTen
* 37" on SUN SparcStation 2
* 31" on IBM Risc 6000-320
* 17" on HP 725/50
* 15.5" on SGI-INDY PC
* 12.6" on IBM Risc 6000-560
* 12.3" on SUN Sparc 20
* 10.4" on SGI-INDY R4400
* 7.5" on HP 735/99
for the complete processing of a 512x2048 data points data-set, including :
-exponential windowing in F2, shifted sine bell in F1.
-hypercomplex FT with zero-filling in F1 and phasing in both axes.
-cubic spline baseline correction through 5 points in F2.
*Post-processing filters; modulus, real part extraction, absolute value,
smoothing, etc...
*Complete baseline correction module (linear, cubic spline correction, and a
comprehensive polynomial module with automatic peak detection).
*Extraction of sub-spectra for local processing.
*Simulation of pseudo data-sets. Simulation of 2D NOESY from intensity files
(CORMA type)
*Addition, multiplication of data-sets.
*3D Display in 3D perspective
*Random access to rows, columns, diagonals or projections in 2D mode.
*Random access to planes and rows in any direction or diagonals in 3D mode.
*Multi-windowing facilities : up to 4 active windows in the same time, many
passive windows.
*Fast zoom mode - Fast interactive phasing mode.
*Complete plotting facilities. On screen : 1D plots; 2D : full colours density
maps, zoom, contour plots and stacked plots; 3D : automatic series of 2D plots,
3D graphics. All 1D, 2D and 3D graphics may be sent to a plotter (HP-plotter or
PostScript laser printer). Several plots may be disposed on the same sheet of
paper, and plot files can be stored separately.
*Maximum Entropy deconvolution of Lorentzian line-shape, of J-coupling pattern,
or of any user-provided function.
*Comprehensive Linear Prediction module, including Burg, LP-SVD, forward and
backward methods, etc...
*Integrated processing of NOESY build-up curves, permitting to accurately
evaluate distances.
*Automatic peak-picker in 1D 2D and 3D; Automatic peak integrator in 1D and
2D.
*Line fitting of mixed Lorentzian and Gaussian Line Shapes
*Several file formats are available, notably a FT-NMRreg. compatible format, a
MATLABreg. compatible format, a compressed format, a generic file format,
permitting direct random access, and several others.
*Calibration, integration, peak-picking, etc..
*Fast interaction with the mouse on 2D and 3D
*Help for attribution, on screen and on plotter
*Easily extensible with the control language
*Interactive commands for beginners as well as fast entry mode for advanced
users
*Complete on-line HELP.
*Comprehensive manual.
*Versatile command parser which permits to build macro commands with alias,
tests, local and global variables, loops, graphic interaction and display
capabilities, support for data-bases and associative arrays.
*Graphical interface completely modifiable and definable by the user.
*Possibility to run in interactive mode as well as Batch mode (display less).
*The operating system is fully available from Gifa.
You enter the program by typing gifa on your favourite system (don't
panic, just type EXIT to get out of it!) . The program should give you a short
greeting, the size of the larger data-set, and then respond in the text window
with the prompt :
Gifa>
If every thing as been set up correctly, you should also see two
spectral windows and a horizontal menu bar showing off. To get the graphics,
you should make sure that the environment variable DISPLAY has been correctly
set to point on your current terminal (with the command setenv DISPLAY when
running UNIX). If no X display is available, Gifa will tell you that no graphic
will be available, but will start correctly however.
The other method consists in clicking on a button in the graphic interface.
The graphic interface consists in sets of related commands, arranged in
vertical button boxes, each such boxes being called by clicking the associated
button in the menu bar. Each button box can be opened and closed at will.
The two methods are strictly equivalent, graphic buttons are internally
associated to commands (or list of commands). One day, you will eventually
learn how to do your own buttons.
Graphic windows can be closed at any time, simply by clicking on their
close-box. The 1D window, the 2D contour-plot an the 3D cube window can also be
resized. However in certain cases (resizing a 1D in 2D mode for instance, or
when a complex drawing has been set, or with the 3D display), the image in the
window can be lost for a short time. However the data can never be damaged in
any manner.
Entering
Gifa> HELP
at the prompt level will give you the same message. The command HELP
is here in uppercase, it can actually be entered also in lowercase, or even in
a mixture of upper and lower case letters. In the whole manual, we will use the
rule that every built-in Gifa command is given in uppercase. Macros, however
have to be typed exactly as defined (this is due to the UNIX file-system), they
will always appear in the manual as lower case.
Typing
Gifa> HULP
will give you an error message, since there is no command nor macro
named hulp (at least on the distribution kit)!
The HELP command can also be used with a parameter :
Gifa> HELP primer
Some command may require one or several parameters, you will be prompted for
such parameters, either in graphic dialogue boxes if the command was entered
from a menu, or at the text terminal if the command was typed from it. When
entering the prompt level, it is also possible to put the parameter directly
after the command on the same line.
For instance, let us try the UNIT command which permits to determine the
current unit used on the data-set : PPM, Hertz (as determined form internal
parameters) or Index (ranging from 1 to n in the case of a data-set of length n
), or seconds (for time domain data-sets).
Typing
Gifa> UNIT
will prompt you for the unit to use, just typing return will leave the
value unchanged.
Typing
Gifa> UNIT H
Will set the current unit to Hertz.
On the other hand, clicking on the Unit button, in the Move menu
will pop-up a dialogue box which realises the same thing.
These windows can be closed at any time, re-opening them is performed by
issuing the correct command either from the prompt or from the menus (see
below). Most of these windows can be resized, to the exception of the 2D
density display window which has a fixed size of 512 pixels on each axis. The
colours in these windows can also be modified with a set of colour commands.
All the spectral windows can be FREEZEd, either by typing the command, or by
selecting the entry in the window menu. A frozen window is just a copy of the
current window, just staying here for comparison. It is completely passive, and
can only be closed or iconified if needed, the content cannot be changed
any-more.
The graphic windows are completely optional, and the effect of a command does
not depend on the fact that a graphic window is there or not to display the
effect.
A control box should also appear at start-up in the upper right corner of the
screen. This box holds controls which permits to zoom and adjust the scale of
the display. (If this box disappears, or is absent for any reason, you can make
it appear again with the ZM command (also in the Display menu)).
To zoom the display, simply click on the graphic window with the left and
middle buttons pressed, and draw a rectangle on the display. You can redraw as
many time as wished this rectangle. To zoom into the selected region, simply
click in this rectangle with the left and right button of the mouse. You can
also use the Zoom in and Zoom out buttons in the control box. The
spectrum showed in this box, is the complete spectral region, as was dispayed
when you clicked on the Catch Spectrum button. The white rectangle in
it, shows the current zoom region.
The four little arrows can be used to move around the zoom region. Each arrow
will move the selected region by half the size of the zoom box.
There are five controls to adjust the vertical scale used for displaying the
spectra. The Reset button will reset the display to the default,
(ABSMAX 0 SCALE 1) where the largest peak in the screen in full size.
The four other buttons permit to modify the scale value, raising or lowering
the SCALE context by factor of 2 and of 1.2. See below for more details on
SCALE.
The pop-up menu Dim, can be used to rapidely switch between the 3
working buffers : 1D, 2D and 3D. Finally, the text above the small spectrum is
the current coordinates of the mouse, that you get whenever you click in the
display.
However it is completely optional, and Gifa can work without any menu bar
(even without any graphic window !), or with a completely different graphic
environment.
The environment macros are found in the /usr/local/gifa/macro directory. Some
macros necessitate additional files for interactive processing or for texts,
these files are held into the /usr/local/gifa/macro/gm directory.
If a menu is buried below other windows, clicking on the menu name will pop
the menu again.
If the graphic is missing, try typing the following command env_base.g, which
executes the macro which loads the default menu-bar, from which you can add all
the specific menus.
Some menus are optional or may not be on screen at the same moment, see the
Mode Menu.
Entries followed by 3 dots (...) indicates that some input will be asked to the
user by the command.
The name of the entries in the menus have been chosen to ressemble as closely
as possible to the command name which would realise the same action. This makes
the menus a bit cryptic, but ease the process of learning the commands, which
can then be used to extend the capabilities of the program by writing macros.
These menus implement most of the actions that you will need in regular
processing, however there are many more capabilities in Gifa than the ones
which are available from this standard menu interface.
This button is equivalent to the HELP command without parameter.
This button is equivalent to the HELP command with a parameter.
This button is equivalent to the apropos macro.
This button is equivalent to the READ command.
This button is equivalent to the WRITE command.
This button is equivalent to the SIZE command.
This button is equivalent to the LIST command.
This button is equivalent to the CONFIG command.
This button is equivalent to the QUIT N command.
This button is equivalent to the DISP1D 1 command.
This button is equivalent to the DISP2D 1 command.
By default when entering the program, the DISP1D and DISP2D windows are on but
the CDISP2D window is off. This is because the display in contour plot is
slower than the density display, and it makes little sense to display a FID in
contour plot!
This button is equivalent to the CDISP2D 1 command.
This button is equivalent to the ZM command.
Equivalent to calling the dispcont macro.
This button is equivalent to the FREEZE command.
This button is equivalent to the RZOOM command.
Equivalent to the super1d macro
Equivalent to the super2d macro
All the processing in Gifa is performed in working buffers. There are 3 main
such buffers, the 1D, the 2D and the 3D buffers. The content of the 1D and 2D
buffers is directly visualised in the 1D and 2D graphic windows. Only one such
working area can be selected at a given time, however switching from one buffer
to the other is virtually instantaneous. All commands and actions apply to the
currently selected buffer; processing commands (FT, etc..) as well as display
commands (SCLAE, ZOOM, etc...).
Choosing a working buffer is completely independent from choosing to display
one data space or another.
A independent buffer is available, which can hold indifferently a 1D, 2D or 3D
data-set. This buffer cannot be processed nor visualised, but can be used as an
additional "hand" for processing or comparison.
Put Data ...any processing... Get Data
can be used as a kind of limited undo facility. Note however that
certain commands use the DATA buffer, thus destroying the Get Data
capability.
Equivalent to the SIN command.
Equivalent to the CHSIZE command.
In 3D processing F3 axis always refer to the acquisition axis, F1 refers to the
axis which is stored with the slowest increment, and F2 to the intermediate
axis. Thus, 3D can be seen a series of F2-F3 planes. Note that this definition
depends only on how the data-set was stored, and not on the order of the
incremented delays in the sequence.
Naming of planes is done by giving the axis orthogonal to the plane rather than
the axes in the planes; for instance the planes holding the F1 and F3 planes
are noted F2 planes.
In the case of in-memory processing, commands are very similar to what is done
in 2D.
This control box permits to change its position and size within the window, to
set the orientation of the spectral cube, to change the perspective of the
display, and finally to recompute a display with the Refresh button.
3D processing on file is done by having an input file and an output file,
planes are loaded from the input ile, processed in memory as regular 2D
experiment, and then copied on the output file.
IFTBIS FT
IFT CHSIZE(%*2) FT REAL
or
IFT FTBIS ; better
You should start with a spectrum, phased, baseline corrected, but, ideally,
not apodised. You can do processing from an apodised data-set but you will get
better results if you start over with a spectrum obtained without any
apodisation.
Using this menu, Maximum Entropy can only be applied on a spectrum containing
only positive, absorptive spectra.
For a complete understanding of MaxEnt processing, read that part of the
manual devoted to MaxEnt.
The command also realizes the inverse Fourier transform step which is necessary
to create the pseudo-FID that will be used later on by MaxEnt.
* you don't have a FID on screen
* you don't have realised the previous steps
During the iterations you should check the following values : chi2,
should be as small as possible, chi2 = 1 means the computation is
finished; conv should be as small as possible, a conv larger than
0.1 means that the convergence will never be obtained, probably because the
data-set or the deconvolution function are incorrectly chosen.
To fully understand what is going on here, you are supposed to have read the
manual (no joking :-). Most (all ?) entries here, are supposed to be applied on
untouched (i.e. not apodised) FID.
This menu is built by the env_lin_pred.g macro.
Equivalent to using the BURG command, or the dt->ar ar->dt
size 1 sequence.
Equivalent to using the dt->svd % svd->ar 1 ar->rt 1
rtreflect 1 rt->ar 1 ar->dt size 1 sequence
It is the burg_rev macro
Prompts you for a size on which the spectrum should be recomputed.
Will prompt you for an exact number of line to keep in the spectm.
The other menu entries permit to realise more specific plots. Each
entry add a element to the current plot, and the final entry Page send
this plot to the plotter.
Commands are presented here ordered by functionality, and oriented toward the
command level, in contrast with the previous chapter oriented toward the user
interface. Extending the user interface can be simply done by learning and
using these commands, and the macro language.
As you have seen if you have been through the primer, Gifa is an interactive
program : it waits for commands from the user. Some commands have parameters
some don't; some commands may even have a variable number of parameters
depending on some context value (it is a so-called context-dependent
grammar).
For instance HELP is a regular command, but DISP1D is a context which handles
the parameter describing the state of the graphic 1D window. A good example of
this distinction is given by the context LB which defines the default value
used for exponential broadening, as compared to the related EM command which
actually apply this exponential broadening to the current data-set.
Most of the behaviour of the program depends on these internal parameters
(contexts). Another example of a context is DIM which describes whether we are
working with 1D 2D or 3D data-sets .
For instance, type :
Gifa> DIM 1 ; switches to 1D NMR
Gifa> size ; lists basic parameters
Gifa> DIM 2 ; switches to 2D NMR
Gifa> size ; lists basic parameters
( characters following the semicolon are comments )
The effect of the command size (which lists the basic parameters of the
current data-set) depends on the state of the context DIM.
Most contexts have associated Gifa variables which permits to evaluate its
value during macro execution. See the chapter on variables for details.
Commands can be issued alone, in which case, the command will prompt you for
all the values it needs, proposing default ones. Default values may be kept by
hitting the <enter> key, or may be changed by typing new ones. You can
also type the parameters on the same line than the command; the effect is then
immediate. When using in-line entry, the different item will be separated by
blanks.
Several commands can even be typed on a single line, separated with a blank.
For instance to get back to 2D display mode, you could either type :
Gifa> DIM 2
Gifa> size
or, on a single line :
Gifa> DIM 2 size
The effect would be the same. The only difference is that the graphic
is only refreshed at the end of the command line; this permits to link together
related commands, without slowing down the process by useless display
(irrelevant in the present example).
* Some commands prompt for trivial parameters : file name (READ), integer
number (DIM with parameter which can be 1, 2 or 3), etc.. Little to say about
theese.
* Some commands promt for spectral coordinates. Coordinates are always prompted
as INDEX (this could be easily changed in the source, to pompt in the current
UNIT), and can be entered in any unit : integer values (terminated or not with
a i) for indexes; in Hertz by terminating with a h; in ppm with the p suffix;
and finaly in seconds with the s suffix. e.g.
zoom 1 10 10 200 300
zoom 1 10i 10i 200i 300i
zoom 1 8.3p 8.3p 7.5p 6.5p
zoom 1 6000h 6000h 4500h 4000h
or even mixed :
zoom 1 10 10i 7.5p 4000h
actually might defined the same zoom window (values are given as an
example)
* Some commands prompt for a direction on the current data-set (for 2D and 3D),
in which case, axes are called : F1, F2, (F3 if in 3D), sometime combinaisons
are meaningfull : F12, (F13, F23, F123). In 2D, F2 is the classical dimension
(associated to t2) and F1 is the non classical (t2). In 3D, F3 is the classical
dimension, F1 is the slowest non-classical and F2 is the intermediate one.
* Some commands ask for a parameter per spectral axis, in which case, F1 is
always prompted first, then F2 (and finally F3).
* Some commands ask for a list of parameters, and the list has no predefined
length : BCORR 1 (which ask for coordinates), FORMBOX which asks for field
definitions, etc.. In which case, the list has to be terminated by some
caracter. It will be 0 whenever integer values or coordinates are awaited, and
it will be a star "*" whenever strings are awaited.
* Some commands prompt for dimension (CX, CY). All dimensions are in
centimeters.
So, to process a data-set with Gifa, you generally load it in memory, do most
of your processing, plot it, and optionally store the processed data on a file
for later processing. This permits a very fast processing on small every-day
data-sets. When a larger data-set necessitates to work on a file, then a set of
specialised commands permits to use all the regular commands on file.
Gifa presents several buffers to store the data. Three working buffers are
available for respectively 1D, 2D and 3D. These 3 buffers are independent for
regular processing. The buffer on which you are working depends on the value of
the context DIM which takes the value 1, 2 or 3.
All the commands refers to the current buffer (as selected by DIM). Depending
on the value of DIM, some commands may ask for a varying number of parameters (
1 per dimensions ) others may ask for direction, in which case the direction is
selected with the syntax Fx. In 2D : F2 for the acquisition dimension, F1 for
the other and F12 for both. In 3D : F3 for the acquisition dimension, F1 for
the slowest dimension, F2 for the intermediate dimension, and F12, F13, F23 and
F123 for combinations. When a command requires a parameter for each dimension,
F1 parameters will be entered first, then F2 and finally F3 (if in 3D), this
rule is general, with the exception of graphical commands which will ask for X
and then Y parameters.
Several file format are available directly from within the program (plus the
utilities permitting to translate from other file formats). The following
format are available : READ or READC (same command) for the standard Gifa
cached format, the file is stored in a submatrix format, the parameters are in
text format in the header part of the file (can be viewed with more); READH is
for the ft-nmr format; READL is for NMR1/NMR2 format; READV is for Varian
format. The corresponding write commands are WRITE (or WRITEC) WRITEH and
WRITEL (no WRITEV available so far).
Several other file formats are available : READM and WRITEM reads and writes
text files, formatted as one line per row for a 2D data-set, and a single line
for 1D (not available for 3D data-sets). This format is compatible with the
-ascii format of the MATLAB program. Note however that no parameters are stored
along with the data, and that the text file might have very long line that will
overflow most text editors. READT and WRITET handle data file in generic text
format; on entry per line, with a set of parameters; this permits to import and
export data from Gifa to any other program. READS and WRITES are special file
format stored in compacted text format; this format is totally independent from
the machine used, the file thus stored can be read with the Gifa program
running on any machine. This permits to share NMR files in a network, for
instance a set of different workstations working with NFS on a file server.
This format is in text format and can be safely sent on E-Mail (however you
should take care not to send too large files, and eventually cut your large
files in smaller pieces (size <= 100kbyte)). Finally a READZ WRITEZ format
is available, this format make use of Linear Predictive Coding to compress the
data before storing (J.Magn.Reson. 1991). The WRITEZ command uses the value of
the ORDER context as the length of the Linear Predictive Polynomial (see
chapter on Linear Prediction). The default value of 10 seems to be an optimum
for most NMR data-sets. The compression is performed with no loss of
information, with a compression ratio typically in the 50% - 60%. To be used
only on time domain data-sets.
EM is for exponential broadening. The parameter is the broadening applied in
Hz. Negative values are possible and are for resolution enhancement (associated
with GM). The value used is given by the value in the context LB. In 2D and 3D,
will apply a different exponential apodisation on axis. If you do not wish to
modify on a given axis, enter a null value.
GM is for Gaussian broadening. The parameter is the broadening applied in Hz.
The value used is given by the value in the context GB.
LB and GB are also used in Maximum Entropy processing.
SIN is for a shifted sine-bell. The parameter is the position of the maximum
of sine-bell, varying form 0 (pure cosine) to 0.5 (pure sine). The parameter of
SIN is not kept in a context.
SQSIN is equivalent to SIN but applies a SQuared SINe-bell.
TM is for trapezoidal windowing, and it uses two parameters which are the
coordinates (in points) of the 2 points T1 and T2. The window then goes from
0.0 value at the first point of the data set to 1.0 at T1 and from the 1.0
value at point T2 to 0.0 at the last point plus one of the data set.
In 1D, you can use the sequence :
Gifa> ONE SIN .3 ; or any other apodisation
to visualise the effect of a given filter.
Depending on the spectrometer the data you get should be processed with FT
(complex FT) or RFT (real FT) and eventually pre-processed with REVF. REVF
actually inverses every 2 points over 4. This has the effect to reduce the
time-proportional format used for the seq mode on BRUKER spectrometers (on real
data-sets).
On complex data-set it has the effect to put the zero-frequency on the borders
of the spectrum (after the FT step!).
The Fourier transforms available are :
FT IFT : complex to complex FT and its inverse
RFT IRFT : real to complex FT and its inverse
FTBIS IFTBIS : complex to real FT and its inverse
The complete set of Fourier transform can also be summarised as follow (C
stands for Complex and R for real) :
FIDs Spectra
C ~FT-> C
C <-IFT~ C
R ~RFT-> C
R <-IRFT~ C
C ~FTBIS-> R
C <-IFTBIS~ R
R Does not exist R
The Hilbert transform is simply realised by
IFTBIS FT ; generate a complex data set from a real
IFTBIS PHASE 90 0 FTBIS ; generate the missing imaginary part
IFT FTBIS ; in-place zero filling using Hilbert
; relations
INVF is related to REVF, it inverse every other point in the data set, thus
taking the conjugated value on complex data-sets. Because of this, the
sequences : INVF FT and FT REVERSE are equivalent. On hypercomplex data (2D)
INVF F12 will take the hyperconjugated.
REVERSE simply returns the current spectra left to right.
The type of the data-set (real or complex) is described by the context ITYPE.
For 1D, ITYPE is 0 for real data and 1 for complex data. For 2D, ITYPE can
takes 4 different values : 0 means real; 1 means complex in the F2 direction; 2
means complex in the F1 direction (I know, this is a bit odd but has to make
sense in 1D as well as in 2D); 3 means complex in both directions (so-called
Hypercomplex data type). In 3D 1 means complex in F3, 2 complex in F2, 4
complex in F1 and the corresponding sums depending on the hypercomplex state of
the data-set. For instance, 7 means that the data-set is complex in all axes.
Certain commands require a specific ITYPE value. For instance FT is available
only on complex data sets. An error message will be given if an FT is tried on
a real data-set. Note that if the data is complex, the real part is held in odd
points of the buffer, the imaginary part is held in the even points.
ITYPE is usually handled automatically by the program, so the user does not
need to worry about it. You may however have to change it in certain cases,
typically when loading the data-set the first time, when "playing around" with
the data-set, or when a "bug" happens in the program (highly
unlikely......!?)
HPHASE performs a Hilbert transform in order to create the missing imaginary
part, it thus permits to phase a real spectrum by creating the imaginary part
on the fly. The sequence IFTBIS FT will create a complex spectrum from a real
one (with half the resolution), thus permitting to experiment phase parameters
with the PH command. These phase parameters can then be used on the
corresponding original real spectrum.
An automatic phasing method have been made available in Gifa lately : it is
the APSL method (by A.Heuer, J.Magn.Reson. 91, p241-253 (1991).
This method works by using the symetry of the phased line. A macro have been
written which implements this method, it is called apsl, and works in 1D.
The command MULTDATA, on the other hand, multiplies point by point the content
of the current buffer with the content of the DATA buffer. This permits to
realise convolution product. When applied on complex (respectively
hypercomplex) data-sets, a complex (respectively hypercomplex) product is
realised.
USWA will separate the real and imaginary part of a complex data-set, they
will be displayed on each side of the data-set, changing its ITYPE from complex
to real, thus permitting to examine both part at the same time.
SWA performs the inverse of USWA by taking each side of a real data-set as
real and imaginary parts of a complex data-set. Both SWA and USWA will only
work on data with a size equal to a power of two.
ABS will compute the absolute value of the current real data set.
MODULUS will compute the modulus of the current complex data-set and put the
result in a new real data-set. In 2D the action taken will depend on the value
of ITYPE :
ITYPE=3, the hypercomplex modulus =
r(RR2+RI2+IR2+II2) will be
computed; ITYPE=1, the complex modulus = r(R2+I2) will
be computed along F2; ITYPE=2 is not handle and the data should be modified
with FLOP. In 3D, the modulus will be computed only for fully hypercomplex
data-sets (ITYPE = 7).
PLUS and MINUS will put to zero all the negative points (the positive for
MINUS), thus leaving a purely positive (negative) data-set.
SMOOTH and MEDIAN are after-processing filters. SMOOTH realises a
moving-average smoothing of the data set on a window of variable length,
whereas MEDIAN will perform a median filter on the data-set, by taking the ith
smaller value of the moving window. Both MEDIAN and SMOOTH will change the
data-set size by the number of point of the moving window.
Linear mode will then realise a linear regression through the pivots points
and remove the linear baseline thus found. If only one point is given, then a
horizontal baseline is computed. Spline mode will compute a cubic spline going
through the pivot points and remove it from the data set. Spline mode requires
at least 3 pivot points to work. BCORR proposes as default values for the index
of the pivot points, the last clicked points with the point (the values in the
point stack) command; you can thus use the %% syntax.
A third option is available in BCORR, corresponding to polynomial (and
related) baseline correction. This last option of the baseline correction is
modular. The correction is made in three steps:
*first the signals and the baseline are separated in zones (referred in the
following as the first segmentation), this first segmentation is eventually
helped by a temporary smoothing of the data,
*these zones are then joined together in several sets of related area (referred
in the following as the second segmentation),
*the correction is finally computed.
There are three general parameters :
BLOCBASE : all parameters in point unit are scaled in the ratio of the data set
size to this parameter.
BLCITER : the maximum number of iterations for the whole process
BLCW : when the RMS of the correction is lower than BLCW times the RMS of the
data, the correction is finished.
There are several commands and parameters to configure the correction :
SMOOTH1 : controls the smoothing of data.
0 : no smoothing.
+1 : moving average of the data set on a window of WINMA points.
+10 : hysteresis smoothing of the data set with the value LEVELHYSTE.
SEGM1 : you can choose the way the first segmentation is done as follows .
0 : without.
1 : with a standard deviation algorithm on the data set.
2 : with a standard deviation algorithm on the first derivative of the data
set.
3 : with thresholds on the data set, and on the first and second derivatives.
4 : with a dynamic clusters algorithm.
+10 : with a morphological filtering after the segmentation.
The two parameters used by the standard deviation algorithm are BLCU and BLCV.
You should only use BLCV, for example if you increase BLCV you reduce the
sensibility of detection in order to correct a dispersive line shape.
The threshold option just remain for historical means, the four thresholds are
SDS, SDB, SCB, SCS.
The dynamic clusters algorithm is controlled by four commands :
DCALGO : select the data on which the algorithm operates.
+1 : the algorithm runs on the data set.
+10 : the algorithm runs on the first derivative of the data set.
+100 : the algorithm runs on the second derivative of the data set.
DCDISTANCE :
0 : Norm 1.
1 : Euclidean distance.
DCITER : the maximum number of iterations for the dynamic clusters algorithm.
DCFACTOR : used in a logarithmic scaling of the data set.
Then you can add a morphological filtering either on the baseline selection or
on the signal selection by choosing a number of points with MORPHOB and
MORPHOS.
SEGM2 : this feature is only useful with a polynomial approximation.
0 : without.
1 : interactive.
2 : automatic.
When you choose an interactive second segmentation, you have to choose one or
several areas with the WINDOW command to allow the program to cut the
correction in these areas if needed.
The automatic segmentation uses a window, WINSEGM2, and a threshold,
LEVELSEGM2.
APPROX : the last step is to choose the way the baseline is estimated.
0 : with ITERMA2 times a moving average filtering on a window of size WINMA2.
1 : with a Legendre polynomial approximation of degree DEGRE.
+10 : with a linear interpolation of signal before approximation.
+100 : with an 'elastic effect' that is a rude way to prevent from burying the
weak lines.
There are four special commands that may help you with all these options :
BCORRP0 : restores the initial configuration with a polynomial approximation.
BCORRP1 : enable a configuration with a dynamic clusters segmentation and a
moving average approximation.
BCORRP? : lists the current configuration.
BCORRP : a step by step command to choose a set up.
ADDBASE will remove a given systematic offset on the data-set. The command
prompts you with the last value computed by EVALN.
The context FREQ holds the basic frequency of the spectrometer (in MHz). It is
used to compute ppm from Hz. The context UNIT will tell other commands (POINT,
PKLIST, etc...) to choose the unit used. UNIT can take the values INDEX (number
of channel) HERTZ (based on SPECW and OFFSET), PPM (HERTZ divided by FREQ), and
SECOND(based on SPECW). All other values are invalid.
The size of the created windows with the DISP2D command, is hard-wired and
cannot be modified.
The display is refreshed at only the end of each command line; you can thus
avoid useless display of intermediate computation by putting all the related
commands on one line.
2D are displayed in density mode. The density mode consists of a colour coding
of the value of the points. 64 colours are used, distributed with a constant
interval on positive and negative values. Values around zero are coded in
black.
The display is computed at the end of the command line. Some point may be
skipped when displaying large spectra (typically greater than 4k in 1D, and
larger than 512x512 in 2D) but not for plotting. The vertical size of the
display is defined by the SCALE context. The display depends also on the ITYPE
value; imaginary points are nor displayed.
The scale is computed such that for SCALE=1, the larger point of the spectrum
is full-screen. The value of this larger point is held in the ABSMAX context.
ABSMAX is recomputed at display time whenever the data actually changed,
however it is not recomputed when it is not needed and you can force the
computation of ABSMAX by putting its value to 0. The scale being relative,
ABSMAX can be modified to compute absolute display and plot by forcing its
value to any pre-defined value. This value will not be overridden as long as
the data are not actually changed.
To make this long story short, one can say that the point with the value
ABSMAX/SCALE will be displayed full screen.
SIGN determined, in 2D or 3D whether only positive points, only negative
points or both will displayed.
VHEIGHT is the level to which the zero level will be plotted in 1D display.
Unit is in %, 0 will draw at the bottom of the screen/page, 1 at the top.
Standard value is 0.3 (30%).
CLEAR context determines if each display will be drawn on the top of the
previous one (0 mode) or if the window will be cleared before drawing (1 mode -
default mode).
1 2 3 4 5 6 etc...
with LOGA 1.5 levels will be :
1 1.5 2.25 3.375 5.0625 7.59 etc...
LOGA equal 2 corresponds to the BRUKER standard spacing.
When working full spectrum on large data-set a CDISP2D can take too long, and
you may wish to stop the display, you can always do it by typing a ^C.
Contour plot display are normally in colours, with a colour code equivalent to
the DISP2D mode, however it is possible to switch to a Black and White mode
with the CCOLOR context.
The colour table used by the density mode is built when starting the program.
The presence of a file called .gifacolor is checked first in the working
directory, then in the $HOME directory, then the presence of a file called
gifacolor is checked in /usr/local/gifa/com. If one of these file is found, it
is used as a list of colours to be used by the density display. The file has
one entry per line, first the choice of colours for the pointer, then for the
density mode, starting with the large negative value ending with large positive
values. Each entry is of the form HUE - SATURATION - INTENSITY. If no file is
found, a default set up is used. You will find on the distribution tape a set
of small programs that permits to build such custom colour tables. The standard
colour table, as well as a Black & White colour table are given as
example.
ZOOM is a command which permits to define a zoom window on the data-set in a
non graphical way. It is perfectly possible to define a zoom window, even if
there is no graphic open. Many commands (LINEFIT, PEAK, etc..) only appliy
within the current zoom window
The macro point permits to interact with the data, it is based on MONOPOINT.
With point, the point stack is cleared, and all the clicked points are stored
into the stack. Point is exited by clicking on the data-set with the right-most
button of the mouse.
REFMACRO controls whether display will be done at the end of each command line
within macro. Default value of 0 means no refreshing during macro execution,
which permits much faster execution. If you write interactive macros with
graphical display, you should set REFMACRO to 1. It is often useful to change
REFMACRO several time within a macro.
PEN permits to change the active pen. In HP-GL, this is the number of the pen;
in Postscript, 8 kind of lines are available :
PCOLOR permits to choose a colour when plotting on a colour Postscript
printer. Only the 8 basic Gifa colours are thus available. If you wish to use
more exotic colours, you should edit the plot file itself.
PAGE will send the page to the plotter, and eject the page on a plotter.
PLOTOFFSET is the constant offset on x and y axes which is applied for each
plots. Each plotting command will see the point at coordinates 0;0 as actually
being offsetted from the true 0;0 of the paper with the PLOTOFFSET values.
PLOTOFFSET permits to stack different plots on a single sheet.
TITLE will plot the text following the command at coordinates x=0, y=CY+1.
PLOT? will prompt you for all the parameters currently used for plotting.
All these commands will prompt you for an output parameter, entering *PL (or
*PLOTTER) will output the plot to the plotter. Entering a file-name will create
a file containing the graphic commands for the plotter. If you send several
plot commands to the same file, the following commands will be appended to the
first one in the plot file, thus permitting to make composite plots on a single
sheet. The PAGE command appends the order for page change, send the file to the
plotter port, and makes Gifa "forget" about the plot file, thus plotting again
on that file will erase the file and start a new plot. FORGET will only makes
Gifa forget the file, without sending it to the plotter.
The plot file can also be sent to the plotter by typing at the operating
system level :
$gifaplot plot_file plotter_type ( VMS or UNIX)
with plotter_type being either postscript or HP-GL, depending on the
type of plot file you have created.
When working in postscript, it is very convenient to use a Display Postscript
program to monitor the state of the current plot file.
CX : length in centimetres of a line.
CY : maximum height of a peak in centimetres.
SCALE : scaling applied to the data before plotting, if scale=1 then the
larger peak on the surface will be CY high, if scale>1 then clipping will
occur.
STDY : each new line is offsetted by STDY centimetres in the Y axis.
STSKIP : every STSKIP line of the data-set will be drawn.
STSHIFT : each new line will be shifted right by STSHIFT points, if negative
then the shift is done on the left. (May be fractional).
STSKEW : determines how the horizontal lines are skewed during plot, (if
negative skewed to the left and to the right if positive.
STPL? permits to check all the parameters in one command.
On screen stack plots are currently limited to 256x128 points (larger plots
generally crashes the graphic).
Other buffers are available :
FILTER holds an apodisation function, used with the APPLY FILTER command (only
if the FILTER context is set to 2). In 2D or 3D, F1, F2 and F3 domains will be
sequential in the buffer. You can design an exotic apodisation function, put it
in the FILTER buffer, and use it.
WINDOW is very similar to FILTER as definition, but not as use. WINDOW is the
description of valid points within the data-set (more precisely of
f(1;<[[sigma]]>)). Thus a null value in WINDOW indicate a point with
total uncertainty. WINDOW usually holds 0.0 or 1.0 entries, but any real values
are valid though negative values have little meaning. The command WINDOW
permits to design the WINDOW function; by resting it to 1.0, or by "digging"
holes in it. WINDOW is used by BCORR 3, MAXENT, etc... It is useful also to put
to zero a certain region in a data set (for instance the water curtain in
3D).
Other buffers are available, used by the MaxEnt or the line-fitting package,
see the specific documentations.
PUT loads the given buffer with the current data-set
GET brings back the given buffer as the current data-set
SHOW displays the given information in the 1D window or in the 2D window
APPLY computes the result of the mathematical operation.
SHOW
GET
PUT
The buffer : can be : APPLY
The command SHOW uses, for some option, the value of the context SCOLOR to
determine the colour to be used by the display.
The two commands GET DATA and PUT DATA permit to put aside a data set for a
while and getting back to it very quickly. Note however that the size of the
larger data-set that can be put aside this way is only a fourth of the larger
Gifa data-set.
The PUT FILTER command (to be issued in 1D mode) has a syntax which depends on
the setting of the NCHANNEL context. NCHANNEL describe how many independent
channels will be considered in the filter window. If NCHANNEL is 1, then the
current data-set will become the filter function. If NCHANNEL is greater than
1, then the command will prompt you for which channel to load the filter
function in. This permits to build a 2D or 3D filter function (in which case,
the ith channel corresponds to the ith dimension); or to
build multichannel deconvolution functions for MaxEnt. If the channel index 0
is given, then the action is as in 1D.
The command PUT permits to put the content of one lower dimension buffer into
a higher data-set. You can thus PUT ROW index or PUT COL index in 2D or PUT
PLANE index in 3D :
Gifa> ROW 1 DIM 1 MULT 0.5 DIM 2 PUT ROW 1
will divide by 2 the first row of the data-set.
This design is optimum whenever the data-set is small enough to be kept in
memory without even using the disk (up to 2 Mega (2D : 1Kx2K or 3D :
128x128x128) points on a typical system). The main drawback is that there is a
definitive upper-limit for the size of the data-set. This limit depends on your
system, but is typically 4 Mega (2D : 2Kx2K or 3D : 128x128x256) on most small
to middle work-stations, and 16 Mega (2D : 4Kx4K or 3D : 256x256x256) on larger
machines. This setting can easily be modified by recompiling the program. If
you need to work on a larger data-set, you will have to resort to the cache
memory system, see below.
The state of the program is described by the DIM context which holds either 1,
2 or 3 (standing for 1D 2D and 3D). Each mode is associated to a buffer, thus
there are 3 buffers, for 1D, 2D and 3D. The unselected buffers remains
unchanged during the processing of other buffers. Most of the commands works on
all three modes, however, most of the commands will change their behaviour :
for instance they will need more parameters.
READL, READH, READV, WRITEL, WRITEH, ADD, ADDDATE, SIZE, LIST, ADDBASE, MULT,
DSA, ABS, ONE, ZERO, PLUS, MINUS, ZM, PEAK, INTEG, LINEFIT
The following commands will now take more parameters instead of 1. In this
case, in 2D the first one always refer to the F1 dimension (non-classical
dimension, displayed vertically) and the second to the F2 dimension (classical
dimension, displayed horizontally) ; in 3D the order will be F1, F2, F3.
CHSIZE, GB, LB, EM, GM, SPECW, OFFSET, EXTRACT, ZOOM, EVALN, SMOOTH, MEDIAN
The following commands will prompt you for an extra parameter which will
describe which dimension the process should be applied : F1 means F1 dimension,
F2 means F2 dimension, (in 3D F3 means F3 dimension) and whenever possible
(noted (*) in the present list), combination F12 and F13 F23 F123 in 3D :
FT(*), IFT(*), RFT(*), IRFT(*), FTBIS(*), IFTBIS(*), PHASE, HPHASE, REVF(*),
REVERSE(*), SIN(*), SQSIN(*), SWA(*),TM(*), USWA(*), BCORR
In 3D, the projection along one axis, onto a plane will be computed and put
into the 2D buffer
Gifa> FT F2 FLIP FT F1
will perform the classical complex Fourier Transform (not
hypercomplex). These commands are used when processing phase modulated
data-sets. The sequence :
Gifa> REVF F2 RFT F2 FLIP REVF F1 INVF FT F1 FLOP MODULUS
will perform the computation of a phase-modulated data-set acquired on
a BRUKER spectrometer in sequential mode.
In 3D, you can exchange the imaginary part of the F3 axis with the F1 or F2
axis.
Transposition is not a natural step when working with Gifa, all the processing
can always be performed randomly in any axis.
The transpose process is performed in-place and is very time consuming. On
typical systems it is probably faster not to transpose when processing the data
set. On very small system with very limited physical memory, it may be faster
to use transpose. Try comparing
Gifa> FT F2 FT F1 ; normal processing
to
Gifa> FT F2 TRANSPOSE FT F2 ; transpose during process
on your system
Gifa> plane F2 index
will select the plane holding the F1 and F3 dimension, at coordinates
F2=index.
Plotting planes extracted from a 3D consists simply in selecting planes with
the PLANE command, and plotting them as 2D with PLOT. For instance the simple
macro :
for i = 1 to si_1_3d
plane F1 $i plot *pl page *pl
endfor
would do the job.
You can also check the more developed macro : plot_plane_3d === distributed
with the program.
Buttons are available with the CHECK3D command, which permits to directly
choose some typical points of view (along each axis, along the main 3D diagonal
of the cube). See above the definition of the box in the Graphic User Interface
manual part.
This is performed through a cache-memory system that is implemented into the
Gifa program. This features has the double advantage of permitting the
processing of very large data-set (no limitation in size, as far as the
software is concerned), and to speed-up the processing of the processing of
large data-sets that can be done otherwise in-memory.
With Gifa V4.0 the cache system has been fully rewritten, in order to extend
its capabilities, and reliability. Most of the limitations that were in the
previous version of the cache have been removed. You should also notice a large
speed increase when working on large files. However, the user interface has
been kept as much as possible unchanged, except for the GETC and PUTC commands
which have now a syntax more similar to other Gifa syntax.
In the previous version of the cache (Cache version 1.0, in Gifa 3.x and in
pre-release of Gifa 4.0), the blocks were of a fixed size of 4096 words (16
kbytes) corresponding to 16x16x16 in 3D, to 64x64 in 2D and to 4k in 1D. This
is no longer true, in the present version (Cache version 2.0, since Gifa 4.0),,
the blocks are adapted to the experiment and to the computer. This means that
the block may take any value, optimised for the current hard-ware (typically
4kbytes and 16 kbytes), and that the division of the blocks will depend on the
sizes of the experiment, in order to optimise access speed in all the
spectroscopic dimensions.
The standard format features also a header which holds all the parameters of
the data (such as dimensionality, sizes, spectral widths, etc...). This header
is in text format, and can easily be displayed with a more command. The user
can easily add his own information in the header with the GETHEADER PUTHEADER
commands. The header is of a fixed size equal to the size of a data block. The
cache system can be used with several files in the same time.
Cache memory access is much faster than a disk access. The block structure of
the file speeds up the processing of large data sets. For instance to access 2
successive planes of a 3D, the first plane will be loaded from the disk, and
then the second will be subsequently held into the cache memory. There is no
real limitation of the size that the cache system may handle, however each file
opened with the cache system will use room in the computer memory allowing to
fit several line (plane) of the 2D (3D) experiment.
It is important to note that writing onto the cache is not equivalent to
writing onto disk. There is some mechanism, that will store on disk the content
of modified block when needed; but the content of the file is not warranted
when working through the cache system (this is very different from the WRITE
command). This has no effect when working from within Gifa itself, since all
file access will go through the cache system that will insure the coherence of
the data. However it may have effect in certain cases such as : *accessing the
file from another program (may be another Gifa); *power failure of the
computer; *a bug in Gifa (?).
When needed, it is possible to "flush" the cache and to copy to disk all the
modified blocks with the commands FLUSH and FLUSHCACHE.
Processing data-sets with the cache memory system usually requires using some
macro for scanning through the complete data-set for the operation to be
completed. A set of macros is provided which will permit an efficient and easy
processing (see below).
GETC loads data from the file to the memory; PUTC copies the content of the
memory to the file. Both commands have a similar syntax. They permit to handle
data areas as well as complete lines, planes and cubes. The action taken
depends on the dimensionality of the JOINed file as well as the value of DIM in
the Gifa working context.
dimensionality of the JOINed file :
GETC / PUTC low up
in 1D, where low and up determines the area to load, or
GETC / PUTC axis ... index ... low up ...
in 2D and 3D where the number of axes, indexes and coordinates
depends on the kind of transfer.
See per command manual for the detailed syntax.
The SHOWC command does not actually load the whole data-set in memory for
displaying, plays game with the cache memory. This is why it is a little slower
than the regular display.
This command is used in the super1d and super2d macros which permit to overlay
several display on screen.
FLUSH flushes onto disk the modified data corresponding the currently JOINed
file. FLUSHCACHE will flush all the file currently JOINed.
Each command requires the name of the input file, the name of the output file,
the axis to process and the commands to apply.
process the data row by row or column by column depending on axis (either F1 or
F2). You can also use proc2d in interactive manner, being prompted for each
value. In this latter case, when entering the list of commands, you can use
several lines, finishing the last empty line with a ^D. The commands are
regular Gifa commands, in 1D mode. Macro are valid.
is equivalent to proc2d, but processes the 3D in a plane wise manner. Be
careful that the command you enter will be in 2D mode, and that whatever plane
you choose for the processing of the 3D, the commands will refer to the plane
as F1, F2.
For instance, you can handle a full memory 2D data-set if you do not wish to
do 3D. However, when working in 3D, if you zero-fill a 2D plane extracted from
the 3D, over the protected area size for 2D, you will destroy a part of the 3D
buffer. You will sometime get the question : 'This will overflow the xx buffer,
Ok?' when there is any risk of destroying one of the buffer. This question is
not asked during macro execution, where you are supposed to know what you are
doing.
With the current set-up, the size of the protected 3D area is typically f(1;2)
of the size of the main buffer, the larger protected 2D is f(1;4) of the larger
3D, and the protected 1D is f(1;4) of the larger 2D.
On another hand, certain operations in Gifa need large work storage, and will
use the top of the main buffer for this; thus being incompatible with a large
data-set. These commands are : Maximum Entropy, the linear prediction package,
the automatic baseline correction BCORR 3 and the PUT DATA and GET DATA
operations. However the amount of memory used depends on the command. The
Maximum Entropy and Linear Prediction (but not the BURG, READC and WRITEC
commands which are in-place) will use the f(3;4) of the main buffer so only
f(1;4) will be left for regular processing; this limitation is independent from
the size of the larger data-set to be processed by MaxEnt which is f(1;8) of
the main buffer. The PUT DATA and GET DATA command will use f(1;2) of the main
buffer. The BCOR 3 command will need only f(1;8) on the top of the main buffer.
With all these commands the remaining of the main buffer can be safely used. If
you try one of these command with a too large data-set in the buffer, the
data-set will be corrupted on the overlapping region.
If you feel that the program is too small for your needs, contact the person
who installed the program on your machine, and ask him to install a larger
definition of the program . Definitions for 1 Mega, 4 Mega and 16 Mega are in
the distribution.
SimSpect2D is the basic command, it takes as parameters : first a data-base of
all the spins along with their chemical shift and linewidth; then a list of
intensities in a simple free format, finally a set of controls, determining the
2D modulation and whether the data-set is symmetrical or not. This macro will
generate a 2D FID, that can be then processed as a regular data-set. Be careful
however, that the process can be quite lengthy, if you compute a large region,
with a lot a peaks and a lot of points.
The chemical shift data-base is in dbm format, and can easily be built from
an ascii file with the mkdbppm macro. The intensity file is in ascii, and can
have been generated by any program. For instance, the CROWD program developped
in our laboratory can create NOESY and ROESY simulation from 3D structures and
dynamic information. However the SimSpect2D macro can be used to simulate any
kind of 2D data-sets.
There is an additional macro available : SimSpect2D_form which creates a form
permitting to very simply enter parameters for the SimSpect2D macro.
The Gifa program has a complete capability to detect, integrate, line fit and
manipulate peaks.
This facility is based on an internal structure holding informations on the
last peak detected. This internal structure is called the peak-table. 3 such
peak tables are simultaneously held in the program, respectively for the 1D, 2D
and 3D data sets. Peak tables can be loaded, listed, read and written onto
disk, and peaks can be selectively removed form the table. The commands
relative to the peak table always refer to the current peak table, as defined
with the DIM context.
The peak table is used by the peak-picker module, integrator module, the line
fitting module and the linear prediction module.
One way of doing is by searching for the largest and smallest points in the
data-set with the MAX command, which set the default values for MINIMAX.
However, the smallest value has usually to be reset by the user to a more
realistic value. For instance you can compute the mean level of noise with the
EVALN command, and enter the noise level time a given scalar as the minimum
intensity for a peak :
point ,select an empty area
max evaln %% minimax (4*$noise) % , loads the value
The command PEAK will then find all the local maxima which lies within the
upper and lower bounds in the data-set, and load the peak table. The peak table
will restrict its search to the currently displayed spectrum, thus permitting
to perform a peak-picking on a restraint area of the spectrum by zooming at it.
When using PEAK in 2D and 3D, you will be prompted for a packing radius. Giving
a non-zero value has the effect of "packing" or "linking" all the peaks which
are less than n points apart from each other into the larger one, thus removing
the entries of the smaller ones from the peak output.
The macro pp has been designed to help in this process.
PKLIST i j will list the contents of the current peak table to the
screen from peak i to peak j. Thus PKLIST %% will list
the complete peak table. The origin of the peak table (picker, integrator, LP,
etc...) is given, and all the descriptors of each peak. The unit used for the
coordinates of the peaks depends on the context UNIT. This command can be
successfully with the CONNECT - DISCONNECT commands to generate listings.
The commands SHOWPEAKS and PLOTPEAKS display respectively on the screen or on
the current plot, the content of the peak table. SHOWPEAK permits to highlight
a specific peak in the peak table, by drawing a cross. SHOWPEAKS and SHOWPEAK
uses the current definition of SCOLOR.
PKSELECT permits to select specific entry into the peak table. Non selected
entry will be lost. PKCLEAR removes all entries from the peak table. It is thus
equivalent to PKSELECT 0. On the other hand PKRM permits to remove a
single entry in the current peak table.
PKCLEAN permits to remove entries in the peak table with intensities smaller
than a given value. PKRESET reload the intensities in the peak-table, as found
on the data-set.
POINT->PK adds the content of the point stack (see above) to the current
peak table. This permits to interactively add missing peaks in the peak
table.
SETPEAK is another command for modifying the content of the peak table : the
coordinates of a peak can be manually entered with this command.
The amoeba is determined by three criteria, with four parameters associated :
RATIO, SLOPE, THRESHOLD, and RADIUS. The first criterion that will trigger will
determine where the extension of the amoeba should stop.
RATIO triggers when the ratio between the largest point in the peak and the
current evaluated point gets below RATIO.
SLOPE triggers when the slope changes. A value of 0 will be triggered anytime
the slope changes from negative to positive (thus starting to climb on another
peak); a larger value will permit more freedom on the slope.
THRESHOLD will trigger whenever the evaluated point is below the (absolute)
value of THRESHOLD.
RADIUS determines the maximum extent of the amoeba from the central peak.
When using the integrator, the noise level and the systematic offset of the
surface should have been evaluated with the EVALN commands
There are two additional contexts that determine fully the PARIS module :
SIGN_PEAK tells the peak-picker if the peaks are to be found either as positive
peaks or as negatives peaks. ZERO_QU tells the integrator that the amoebae
should be computed on the absolute value of the surface, (but the integrator
will still work on the normal surface). This is very useful when working on
NOESY spectra where there is zero-quantum coherences signals (the integration
of which is zero).
The SHOW AMOEBA command shows the amoebae that have been found at the last
integration step.
The MSKINTEG will integrate the peaks as define by the current peak table and
mask (amoeba) matrix. This command permits to integrate several experiments
using the same amoeba definition. The amoeba matrix can moved around with the
PUT and GET commands. The 2 macro mskread and mskwrite permit
to read and write directly amoeba matrices along with peak tables.
The amoeba can also be modified with the MODIFY_AMOEBA command which permits
to set a given pixel to a given peak, or to remove it from the amoeba
definition.
mskread and mskwrite are 2 macros meant for storing on file and retreiving the
current peak and amoeba definitions.
Where Y is the recomputed spectrum, y is the current spectrum and [[sigma]] is
the variance of the noise, estimated from the NOISE command. Thus a correct fit
corresponds to a final equal to 1.
Results can be examine with the standard commands SHOW LINEFIT or SHOWPEAKS. As
an addition, macros called show_fit and plot_fit are provided, which permit to
display/plot a composite display of the current data set with the fitted lines
superimposed on the spectrum.
The command :
put data get linefit mult -1 adddata
Permits to obtain the residue (that part of the data which is not
fitted), the command
get data
returning to the current data-set.
The LINEFITD (linefit detailed) command permits to finely select which peak and
which parameters will be fitted, and which will not, for instance fitting only
the amplitude in a series of relaxation measurements, or fitting only the
frequencies in a titration.
PRESENTATION
The Gifa program is a multi-purpose NMR program. It is designed for the
processing, the display and the analysis of 1D, 2D & 3D NMR data-sets.
Classical processing includes :
*Full capabilities in 1D, 2D and 3D processing.
Graphic capabilities
*2D Displays in -density mode -contour mode -stacked plot mode
Special processing includes :
*Maximum Entropy processing of 1D as well as 2D data-sets. A generalised
iteration scheme, (Gifa stands for Generalised Iterative Fixed-point Algorithm)
as well as more classical algorithms : Gull & Daniel, conjugated
gradients.
A Complete set of Macro :
*A complete graphic user interface written in the macro language
The User Interface is characterised by :
*Most command are available through a flexible graphic interface
FIRST CONTACT WITH THE PROGRAM.
Entering the program
First insure that the program have been correctly installed from the
distribution kit, contact your system manager or your local expert.
Principe of operation
There are two ways of issuing commands to Gifa. The first one is to type
commands directly in the text window, at the prompt level. Such commands are of
two kinds, built-in commands, directly executed by Gifa, and macros, which are
command files written in the Gifa command language. From the user level, there
is little difference between the two kind of commands.
Entering Parameters
You are now ready to enter your first command. If you hit the About
GIFA button in the File button box (the box will show off if you
hit the File button in the menu bar), you will get a short
introduction.
BASIC GRAPHIC USER INTERFACE
The standard graphic interface is characterised by several graphic windows and
controls used for the display of the spectral data, and by a set of menus,
button boxes, and dialogue boxes used for graphic user interaction.
Display and Zooming
Several graphic windows can be opened at the same time within Gifa. By
default, when entering the program, a 1D display and a 2D density display
window are opened. Several other graphic windows can also be opened (see
CDISP2D, DISP3D, FREEZE). The data held in the working buffers are shown in
these windows.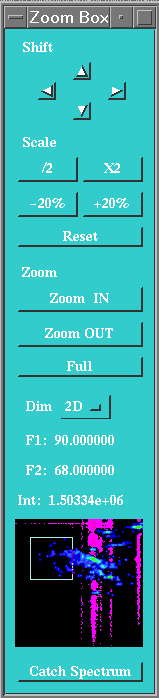
Menu Set-up
The basic-menu graphic user interface is installed in the standard start-up
procedure. It is installed whenever the startup.g macro is executed. This macro
loads the basic environment, and the 1D and 2D processing menus.
Basic environment
The following menus are usually present whenever the graphic interface is
activated, they are independent from the kind or work currently in progress.
File menu
This menu holds the basic commands for getting informations on the program,
reading and writing data-sets and quitting the program.
About GIFA
Will print introduction text, giving the some ideas on the Gifa program, as
well as some hints on how getting more information or help.
Help...
Will prompt you for a command/macro name and will display the help associated
with that command/macro.
APropos...
Will prompt you for a word, and will show the name of all the commands/macros
that have this word in their help file (case insensitive). Try for instance
Fourier or display
Documentation...
This button launch the Netscape program, with an hyper-text version of the
current manual.
Read...
Will prompt you for the name of the file to be loaded into memory. The program
will switch to 1D, 2D or 3D depending on the data-set itself. File should be in
the standard Gifa format.
Write...
Will prompt you for the name of the file to be written from the data held into
memory. The data-set will be created depending on the DIM currently selected
(1D, 2D or 3D).
Read all format - Write all format
Are equivalent to Read and Write but a selection box permits to
choose among all the supported Gifa file formats. For details, see below in the
manual at the READx command
Convert from UXNMR
This button calls the shell script ux2cach.sh which permits to convert from a
Bruker/UXNMR file format (acquisition data, not the processed data) to the
standard Gifa file format. The command first prompt for the directory in which
the data resides (for instance /u/data/guest/nmr/your_exp/1 ) and then to the
Gifa file format. It will then ask for information which cannot be found in the
Bruker files.
File name
Will give you the name of the last read or written file with the READ or WRITE
commands (or the Read... or Write... button).
Size of data
Will give you the detail on the dimension, the sizes, the spectral widths,
etc... of the current data-set.
More details
Will give you the even more details the current data-set, such as the zoom
window, the display values, etc...
Config
Will give all the details on the internal configuration of the program. You
will have information on the current version number and the licence of the
program, as well as the value of the basic parameters. Some values are
hard-wired in the program at compile time, such as the size of the working
buffer, or the number of user variables; some values may be modified by the
user such as the kind of plotter currently in use (see below).
bug report
Permits to edit and mail to me (mad@cbs.univ-montp1.fr) a short bug report.
Quit GIFA
Will quit the Gifa program loosing all data in memory not saved with a WRITE
command, and removing the journaling file. If you want to keep the journaling
file (a file called gifa.log in your home directory, which holds all the
command issued during the Gifa session), you will have to type in the QUIT
command directly.
Mode menu
This menu permit to choose between different interface, already prepared in
the standard distribution. Some button will completely change the interface,
some, noted with 3 leading points, will add a menu at the end of the menu-bar
Proc 1D
Will activate a graphic interface suitable for processing and displaying 1D
data-sets only.
Proc 2D
Will activate a graphic interface suitable for processing and displaying 2D and
1D data-sets.
Proc 3D
Will activate a graphic interface suitable for processing and displaying 3D, 2D
and 1D data-sets.
... Advanced proc
Will add a menu permitting extended processing, such as baseline correction,
water suppression, etc...
...MaxEnt
Will add a menu permitting to realise MaxEnt processing of the data-sets.
...Linear Prediction
Will add a menu permitting Linear Prediction processing.
... Plot
Will add a menu permitting all commands associated to plots.
... Peak
Will add a menu permitting all commands associated to Peak-picking and Line
fitting.
... Unix
Will add a menu permitting Unix-like commands.
Display menu
This menu holds the commands permitting to interact with the display.
Disp1D on
Will switch on the graphic window associated to the 1D buffer. Has no effect if
the window is already displayed.
Disp2D on
Will switch on the graphic window associated to the 2D buffer, displaying the
density plot. Has no effect if the window is already displayed.
CDisp2D on
Will switch on the graphic window associated to the 2D buffer, displaying the
contour plot. Has no effect if the window is already displayed.
Zm (Zoom Control)
This button will enter a Zoom mode, were you can define a zoom window, zoom in
and out, and modify the scale of display. The four little arrows permit to move
the zoom window around without modifying its size.
Display Control
This button will launch a form which permits to control other display
parameters such as the state of the contour display window, the display of the
negative regions, the number of levels and the spacing algorithm.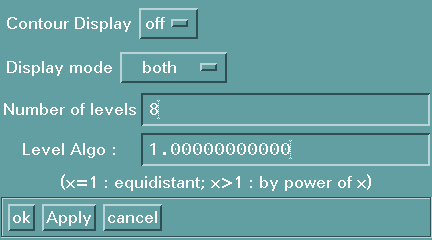
Freeze
This button will permit you to "freeze" a currently opened window, by
duplicating it. It is a bit like taking a snapshot of the window, letting it on
screen as long as you wish. There is no way of altering a FREEZEd window but
closing it.
store zoom
Clicking this will memorize the current zoom window, it will be possible, at
any later time to click on the back to stored zoom to go back to that
window.
back to stored zoom
Jumps back to the last stored zoom window
Rzoom
When in zoom mode, this button will jump to symmetrical zoom region, relative
to the diagonal.
super1D
Creates a form box which permits to display up to 2 additional 1D spectra on
the top of the current 1D sprectum held in memory
super2D
Creates a form box which permits to display up to 2 additional 2D spectra on
the top of the current 2D sprectum held in memory, and displayed on the contour
(CDisp2D on) window.
Move menu
This menu holds all the commands permitting to navigate in the different mode
of the program, or to move data between the different buffers.
dim 1 - dim 2 - dim 3
Switches the active working buffer. Respectively to 1D, 2D and 3D. These
buttons are equivalent to the DIM x command
Put Data
Loads the DATA buffer with the contents of the currently active buffer.
Equivalent to the PUT DATA command.
Get Data
Brings back the content of the DATA buffer into the working buffer. Equivalent
to the GET DATA command. The sequence
AddData
Adds the content of the DATA buffer with the content of the working buffer, and
put the result into the working buffer. To make a difference spectra, simply
multiply the working buffer by -1 (mult -1) before adding. Equivalent
to the ADDDATA command.
Point
Enter an interactive mode, where the coordinates of the cursor are displayed.
Click into the text box to exit this mode. Equivalent to the point
macro.
Unit...
Permits to choose the unit in which the coordinates of the cursor are displayed
during the Point mode. Unit can be either Index, Hertz or ppm.
Equivalent to the UNIT command.
Calib...
Starts a macro (calib) which permits to calibrate the current spectrum, by
clicking on the reference peak, then entering its coordinates.
Select Row
Starts the macro rowint, which permits to interactively select rows
(F2 sections) in the 2D window and display them in the 1D window. You exit this
mode by clicking on the right mouse button. The last selected row is copied
into the 1D working buffer.
Select Col
Starts the macro colint, which permits to interactively select columns
(F1 sections) in the 2D window and display them in the 1D window. You exit this
mode by clicking on the right mouse button. The last selected column is copied
into the 1D working buffer.
Row...
Permits to copy a 2D row, selected by its index, into the 1D working buffer.
Equivalent to the ROW command.
Col...
Permits to copy a 2D column, selected by its index, into the 1D working buffer.
Equivalent to the COL command.
Processing
These menus permit to perform most of the regular processing to be performed
on simple NMR spectra.
Apodisation menu
This menu contains all the operation which are to be applied before the
Fourier transform step and is common to all processing modules (1D,2D and 3D),
since the apodisation step is the same in all cases. It is installed with the
env_proc1d.g macro, along with the Proc 1D menu.
Em...
Apply an exponential apodisation to the current data-set. The value of the
exponential is in Hertz. There is one parameter for each axis in the data-set
(1 in 1D, 2 in 2D, 3 in 3D). Equivalent to the EM command.
Gm...
Apply an Gaussian apodisation to the current data-set. The value of the
Gaussian is in Hertz. There is one parameter for each axis in the data-set (1
in 1D, 2 in 2D, 3 in 3D). Equivalent to the GM command.
Sin...
Apply a sine-bell apodisation to the current data-set. The first (and only in
1D) parameter is a value ranging from 0.0 to 0.5; the value of the parameter
corresponds to the position of the maximum of the filter in your window, thus
0.0 is pure cosine, 0.5 is pure sine, all intermediate values are possible. In
2D or 3D you will be prompted for the axis on which the apodisation should be
applied.
SqSin...
As the Sin button but applies a squared sine-bell. Equivalent to the
SQSIN command.
Tm...
Applies a trapezoidal apodisation. You will be prompted for two parameters, the
built function starts from 0.0, raises up to the index entered as the first
parameters where it reaches 1.0, the decay from 1.0 to 0.0 starts a the index
entered as the second parameter up to the end of the data-set. In 2D or 3D you
will be prompted for the axis on which the apodisation should be applied.
Equivalent to the TM command.
Correct 1st point
Correct the first point of the current data set for the quantification bias
introduced by the sampling. This is typical correction on 2D-3D data-sets. It
partially removes the t1-ridges that are commonly seen. It has less
interest in 1D.
ChSize...
Permits to change the size of the current data-set. The operation (reduction or
increase) will be realised at once on the data-set. Increasing the size will be
realised by adding zeros at the end of the buffer. You will be prompted for a
dimension per axis in the data-set.
Proc 1D menu
This menu, installed with the env_proc1d.g macro, holds the commands for 1D
processing.
ZeroFill
Extend the size of the current data-set to a size equal to the next power of
two.
ft_Seq
Realises the Fourier transform suitable for a data set acquired in
sequential mode : real and imaginary part alternatively sampled.
Equivalent to the ft_seq macro or to the REVF RFT command.
ft_Sim
Realises the Fourier transform suitable for a data set acquired in
simultaneous mode : real and imaginary part sampled at the same time
point, and stored alternatively in the buffer. Equivalent to the ft_sim
macro or to the REVF FT command.
Ph
Starts the PH command which permits to interactively phase the current
spectrum. Pops up a control graphic box, which permits to control zeroth and
first order phase corrections, place the pivot (with the middle button of the
mouse), and store the phase correction. Zoom and the Zoom Box controls are
still active.
Auto. Phasing
Computes a phase correction on the current 1D data-set, using the APSL
techniques (A.Heuer J.Magn.Reson. 91 p241 (1991) ). This is equivalent to
calling the apsl macro.
Redo Phase
Apply to the data-set the last phase correction used. Equivalent to the
PHASE %% command.
Real
Remove the imaginary part of the data-set, thus reducing by two the number of
buffer points. Equivalent to the REAL command.
Modulus
Compute the modulus of the spectrum from the real and imaginary parts.
Equivalent to the MODULUS command.
Proc 2D menu
This menu, installed with the env_proc2d.g macro, holds the commands for 2D
processing. The F1 axis always refers to the non-classical, vertical axis of
the 2D, and the F2 axis to the classical, horizontal axis. Some commands are
specific to F1 or F2 processing, some commandsprompt for the axis to process,
in which case you have to enter either F1, F2 or F12 if you want a processing
on both axes.
Easy 2D
This button launches a form which simplifies the Fourier transform process. You
select the sizes, the apodisation functions, the Fourier, phasing and Baseline
correction steps. The set-up can be used for direct computation as well as to
store a macro that will realise the process in a latter stage.
ZeroFill F1 - ZeroFill F2
Increases the size of the dataset in the F1 or F2 domain up to the next power
of 2.
Burg 2D
This button permits to use the burg Linear prediction method for extending the
data-set. You can see it like a sophisticated zero-filling function.
ft_Seq (F2)
Realises the Fourier transform in F2 suitable for a data set acquired in
sequential mode : real and imaginary part alternatively sampled.
Equivalent to the ft_seq macro or to the REVF F2 RFT F2
command.
ft_Sim (F2)
Realises the Fourier transform in F2 suitable for a data set acquired in
simultaneous mode : real and imaginary part sampled same time point, and
stored alternatively in the buffer. Equivalent to the ft_sim macro or
to the REVF F2 FT F2 command.
ft_phase_modu (F1)
Realises the Fourier transform in F1 suitable for a data set acquired in phase
modulation mode. Equivalent to the ft_phase_modu macro or to
the FLIP REVF F1 FT F1 FLOP command.
ft_tppi (F1)
Realises the Fourier transform in F1 suitable for a data set acquired in TPPI
mode. Equivalent to the ft_tppi macro or to the RFT F1
command.
ft_sh (F1)
Realises the Fourier transform in F1 suitable for a data set acquired in
Hypercomplex (States-Haberkorn) mode. Equivalent to the ft_sh
macro or to the REVF F1 FT F1 command.
ft_sh_tppi (F1)
Realises the Fourier transform in F1 suitable for a data set acquired in TPPI
Hypercomplex (TPPI States-Haberkorn) mode. Equivalent to the
ft_sh_tppi macro or to the FT F1 command.
ft_n+p (F1)
Realises the Fourier transform in F1 suitable for a data set acquired in
echo-antiecho mode, with alternated gradients. Equivalent to the
ft_n+p macro.
Ph in f1 (ph2dc)
Starts the macro ph2dc which permits to interactively phase a 2D
spectrum in F1. You first enter a selection mode, where you click on the 2D
window with the left button to visualise the columns, you select them with the
middle button, the selected columns are added together. You exit this mode with
the right button. You then enter the PH command with a composite 1D
spectrum obtained from the selected columns. The phase obtained on this
spectrum is then applied on the complete 2D.
Ph in f2 (ph2dr)
Equivalent to ph2dc for the F2 domain.
Redo Phase F1
Apply again the last phase correction in the F1 domain. Equivalent to the
PHASE % % F1 command
Redo Phase F2
Apply again the last phase correction in the F2 domain. Equivalent to the
PHASE % % F2 command
Real F12
Remove the imaginary parts of the data-set along each axis, thus reducing the
total size of the data-set by four.
Modulus
Computes the modulus spectrum from the real and imaginary parts. Equivalent to
the MODULUS command.
Proj F1
Computes the projection of the 2D dataset along the F1 axis, onto the F2 axis.
The command will promt you wether you want a "Skyline" (keep the highest
points) or a "Mean" (sum all points) projection. Equivalent to the PROJ F1
command.
Proj F2
Computes the projection of the 2D dataset along the F2 axis, onto the F1 axis.
The command will promt you wether you want a "Skyline" (keep the highest
points) or a "Mean" (sum all points) projection. Equivalent to the PROJ F2
command.
3D menus
When 3D processing has been chosen, either with the env_proc23d.g macro, or the
Proc 3D option in the Mode menu, 3 additionnal menus
are inserted in the menu bar. The first menu is for processing 3D data-sets
which can be held into the central memory, the second is specific of 3D
display, and the third one is for processing and displaying file which are too
big for fitting into the central memory, and thus have to be processed on
file.
Proc 3D
This menu holds most of the commands for processing 3D experiments in-memory,
as it is done for 2Ds. This can be done only for not too big data-sets. In the
contrary case, one has to resort to on-file processing (see below). The Gifa
central memory is determined at compile time, and several versions of the
program, corresponding to differesnt size of the memory, should be available on
your machine, check the installation manual.
Largest Set
This button simply call the largest_3d macro which gives you example
of data-sets which fit into the memory.
ZeroFill F1
Simply realizes extension of the size in F1, to the next power of two. Done
with the CHSIZE command.
ZeroFill F2
Realizes extension of the size in F2, to the next power of two. Done with the
CHSIZE command.
ZeroFill F3
Realizes extension of the size in F2, to the next power of two. Done with the
CHSIZE command.
Burg3d
Performs the burg extension of the data-set in F1 or F2 using the
burg3d macro command.
ft_Seq (F3)
Performs in the F3 (acquisition) axis, the Fourier transform of a data-set
acquired in sequential mode.
ft_Sim (F3)
Performs in the F3 (acquisition) axis, the Fourier transform of a data-set
acquired in simultaneous (complex) mode.
ft_sh_tppi (F2)
ft_tppi (F1)
ft_sh (F1)
ft_sh_tppi (F1)
Performs in the F2 or F1 axis, the Fourier transform suitable for data-sets
acquired with different protocols, see the Proc 2D menu for details.
Phase
Permits to apply a phase correction in a given spectral axis. Equivalent to the
PHASE command.
Real...
Permits to throw away the imaginary part associated with a given axis (F1, F2
or F3). Equivalent to the REAL command
Real F123
Permits to throw away all the imaginary part associated to a fully hypercomplex
data-set. Equivalent to the REAL F123 command.
Modulus
Computes the modulus of fully hypercomplex data-set. Equivalent to the
MODULUS command.
Display 3D
This menu holds all the functions for displaying the 3D data-set held into
memory
disp3d on
Opens the 3D display window, equivalent to the DISP3D 1 command.
parameters...
Opens a form for setting 3D display parameters. actually calls the
disp3d_form macro.
control box
Opens a control box which permit to interactively chose the view point of the
3D display. Equivalent to the CHECK3D 1 command.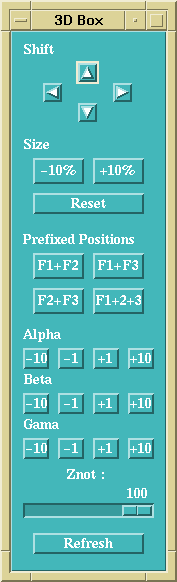
refresh
In contrast with the other windows, the 3D display is not automatically
refreshed whenever a display parameter changes. This has been chosen because of
the time that this refresh might take. Clicking on this button will start the
3D display. Equivalent to the REF3D command, or to the Refresh
buttom on the CHECK3D control box.
choose zoom...
This button starts the zoom3di macro which permits to interactively
choose a zoom window for the current 3D display.
plane...
Calls the PLANE command, which loads the 2D buffer with a plane
extracted from the 3D.
diag...
Calls the DIAG command, which loads the 2D buffer with a diagonal
plane extracted from the 3D
vertint
Starts the vertint macro which permits to interactively examinate 1D
spectra by clicking on a orthogonal 2D plane.
planeint
Starts the planeint macro which permits to interactively examinate
planes by clicking on a orthogonal 1D spectrum
Proj F3 ...
Compute the projection plane along the corresponding axis. You will get
prompted for the projection mode Skyline / Mean.
3D on file
When a data-set is too big to be processed in-memory, it is possible to realize
the processing on-file. This is done with a special set-up in Gifa, the "cache"
system, which permits to access files in an optimal fashion, (see below the
Working On File rather than In Memory chapter).
Join data-set...
This entry permits to realize the first operation to be done, namely connecting
to the input file. This is equivalent to the JOIN command. Once a
data-set has been JOINed, it is possible to display internal parameters, load
1D or 2D from it, or even load a 3D region form it. Note that you can issue
that command to several files, thus being JOINed to more than one file. However
all the command will refer to the last JOINed file.
Dataset parameters
That button simply runs the dataset macro, which displays all the
spectral parameters of the current JOINed data-set. Note that since the header
holding all the parameters is in ASCII format, one can simply make more
filename to see the value of all the parameters.
list all files
This button lists the names of all the files which are currently JOINed.
disjoin
Once a file have been JOINed it must be DISJOINed in order to release
completely the internal memory.
get plane
This menu entry permits to load a plane extracted from the currently JOINed 3D
dataset. For intance it can usefull to load the first F1 or F2 planes to
determines the optimum processing parameters, and phases, before going to full
3D processing. It can used also to look at planes in a processed data-set. It
is equivalent to using the GETC command in DIM 2 mode.
get region
This ones permits to load in the central memory a portion of the currently
JOINed data-set. You will be prmpted for the coordinates fits point (F1, F2,
F3) and then for the coordinates of the last point (F1, F2, F3). It is
equivalent to using the GETC command in DIM 3 mode.
3D proc...
This button creates a form which permits to process the 3D data-set on-file.
One has to enter the name of the input and output files, the axis along which
the processing will be done, and finaly the command line that will be applied
to each plane of the 3D. This command line will be executed in 2D mode on each
of the planes; thus typical 2D command should be given here. The forms actually
calls the proc3d macro.
Proj F3 ...
Compute the projection plane along the corresponding axis. You will get
prompted for the projection mode Skyline / Mean
Optional menus
The following optional menus can be accessed from the Mode menu.
Advanced proc menu
BCorr linear
Apply a linear baseline correction to the current data-set. You have to click
on empty regions of the data-set, from which the baseline correction will be
computed. Equivalent to the point and BCORR 1 commands.
BCorr Spline
Apply a spline baseline correction to the current data-set. You have to click
on empty regions of the data-set, from which the baseline correction will be
computed. Equivalent to the point and BCORR 2 commands.
BCorr Polyn.
Apply an automatic peak detection followed by a polynomial baseline correction.
Equivalent to the BCORR 3 command.
1D Hilbert
Realises the 1D Hilbert transform of the current data-set, that is the current
real data-set is transformed to a complex data-set with a reconstructed
imaginary part. The operation is done without changing the total number of data
points. Is equivalent to
1D invHilb
Realises the 1D inverse Hilbert transform of the current data-set, that is the
current complex data-set is transformed to a real data-set with all the
information of the imaginary part brought back to the real region. The
operation is done without changing the total number of data points. Is
equivalent to
2d Hilbertsxx and 2DinvHilb xx
These menus realises the direct and inverse Hilbert transform of the 2D
data-set.
Remove H2O
Starts the rem_h2o macro which remove substantially solvent signal at
zero frequency in the FID. It does so by removing any slow varying signal in
the FID, using the baseline correction module. Should be applied on the
time-domain data-set, before any other processing (before apodisation or
Fourier transform).
MaxEnt menu
This menu holds some commands than permit to simply process a 1D or 2D
data-set using Maximum Entropy deconvolution. The menus are more or less
organised in the order in which you should use them.
Prepare Data
With this command you first select a small region of the spectrum that you wish
to process. You should not use a complete spectrum, but rather concentrate on
the interest region with this command.
Eval Noise...
Before running the process you should determine the noise quantity present in
the pseudo-FID by zooming, with this command, on the end of FID holding only
noise. If the starting data-set had been apodised, the noise quantity will be
largely under-estimated; if the pseudo-FID is truncated (total decay is not
obtained), the noise quantity will be overestimated. In any case you may want
to correct it with the NOISE command.
Deconvolution fct...
Before running the process you should determine the deconvolution function,
which can be either Lorentzian or Gaussian (other possibility are available
from the command level). Line-width is entered in Hz. The line-shape thus
defined will be removed from the actual spectrum, thus producing sharer lines.
So you should never enter here a line-shape larger than the sharper line
present in the spectrum.
Iterations...
Determines the number of iterations that will be used for the starting
computation.
Start Iteration
Starts the MaxEnt iterations. You are first prompted for the size of the
spectrum to be computed, which should larger or equal to the size of the
pseudo-FID. Never starts the computation if
10 more
Continue the processing for 10 more iterations.
Restart
Start over the processing with the pseudo-FID.
remove background...
When the computation is completely finished, you should remove the background
that MaxEnt always adds to the spectrum, by running this command and zooming in
an empty region of the spectrum. After this operation, the process should be
started over. (10 more is not available any more)
Linear Prediction menu
This menu holds all the entries for realising simple Linear prediction
calculations. (stil experimental - the menu I mean, the commands are fully
operationnal)
Order
Permits to determine the value of ORDER used for all LP operation. Should be
set larger than the number of lines in the spectrum, and smaller than the
data-set size. The speed of LP operations usually depend on this setting.(see
LP documentation). Equivalent to the ORDER command.
make FID Cplx
LP commands work only on Complex FIDs, this button transform a Real FID into a
Complex one by using a Hilbert transform. Equivalent to the command IFTBIS
FT
burg extension
Extend the current FID using the burg method. Can be used to correct truncated
FID. To be used BEFORE any apodisation. Will promt you for the new size.
SVD extension
Is equivalent to the previous button, but uses the SVD method which is somewhat
slower, but more efficient in the case of noisy data. Try this one if burg
failed. Equivalent to using the dt->svd % svd->ar 1 ar->dt
size 1 sequence
stable SVD extension
Equivalent to the precedent, but even slower. That one should succeed in all
cases. To be used when the previous one failed (typically by transforming your
FID into "trumpets" (increasing exponentially towards the end))
burg 1st points
Uses the burg method to build missing first points. Can be used for instance
after a lshift to correct for acoustic ringing.
SVD 1st points
Equivalent to the previous, but with the SVD method. It is the svd_rev macro.
burg spectrum
Computes the "urg spectrum" (also called mem1) i.e. an estimate of the power
spectrum, based on the burg analysis of the FID.
SVD analysis
Performs a complete SVD analysis of the current FID. Results are put into the
pklist
FAB-SVD analysis
The same as previous, but uses the Forward & Backward method. Meant
to be used on very bas signal/noise cases.
Cadzow proc.
Applies the Cadzow procedure to clean up the current FID. Calls the cadow macro.
PKlist
Actually prints the content of the peak table. Equivalent to the PKLIST %%
command.
Plot menu
This menu holds all the entries for realising simple plots.
Easy plot
This button launches a from which permits to determine all the parameters for a
standard plot, with axes, grid, choice of colours, etc...
plot?...
Starts the macro plot? which prompts you for some basic parameters associated
to plotting : the kind of plotter (postscript or HP-GL), the size of the plot
in centimetres, the offset of the plot from the lower left corner of the page,
the vertical offset of 1D plots, the rotate state and the parameters for
spectral axes. Can used to pre-set the values in Easy Plot
Screen Dump
Adds the current display (1D or 2D) to the plot.
Title
Adds a title to the current page. When entering more than one word, put the
complete string into quotes (') or double quotes (").
Plotaxis F1 - Plotaxis F2
Add an labelled axes on the F1 (y) or F2 (x) direction, as defined with the
Plot? command.
Grid F1 - Grid F2
Add a grid in the F1 or F2 domain according to the definition of the axis from
Plot? command.
Page
Send the current plot to the plotter. On certain platform, the plot file is
displayed on screen before being actually sent.
Cindy...
Makes a plot file which can then be read with the NMR analysis Cindy program
(see ftp://tome.cbs.univ-montp1.fr/pub/Cindy). Equivalent to the FPLOT
command.
Peaks menu
This menu holds certain facilities for realising peak detection, peak
integration and line fitting. All the information is held into an internal
table which can be displayed at any time with the PKLIST button.
eval noise
Asks you to zoom on an empty region, and evaluates noise on the selected
region. Uses the EVALN command..
Peak pick (pp)
Enter the macro pp which performs a simple peak-picking. You should
have evaluated the noise level with the previous button Then zoom on the region
to be peak-picked. The peak threshold is evaluated from the noise level.
add a pick
Permits to add a peak into the table by clicking on the data-set. Equivalent to
the point POINT-
>PK
command.
rem a pick
Calls the pkrmi macro which permits to interactively remove a peak on
the data-set.
pkclear
Equivalent to the PKCLEAR command, which removes all peak entries.
Integ
Performs a numeric integration of the data-set by computing masks around each
peak in the peak table, and integrating over these masks. Equivalent to the
INTEG %% command.
ShowPeaks
Display graphically the peaks on the current display. Equivalent to the
SHOWPEAKS command.
PlotPeaks
Equivalent to ShowPeaks but on a plot file. Equivalent to the
PLOTPEAKS command
Show Amoeba
Display the masks used by the last INTEG command. Available only in 2D.
Equivalent to the SHOW AMOEBA command.
linefit
Calls the generic line fitter. You are prompted for the line-shape to fit
(Lorentz or Gauss). Equivalent to the LINEFIT command.
show linefit
Equivalent to the SHOW LINEFIT command. Display the content of the curent peak
table as a simulated spectrum. Can be used after LINEFIT, but also at any stage.
show_fit
Graphically displays the content of the peak table (as obtained for instance,
from the last line fitting). Equivalent to the show_fit macro.
residue
Display the residue of the last line-fitting. Data can be restored with the
GET DATA command.
PkList
Lists the content of the internal peak-table. Equivalent to the PKLIST
%% command.
PkRead
Reads the content of the peak table on a text file previously stored with
PkWrite. Equivalent to the PKREAD command.
PkWrite
Writes the content of the peak table on a text file. Equivalent to the
PKWRITE command.
Unix Menu
This button holds some facilities for interacting with the UNIX Operating
System. All the buttons are implemented through the generic SH
command. All entries have equivalent macros or commands available in text mode.
pwd - cd... - more... - ls -l
Realise the Unix counterparts.
Shell
Opens an XTerm with the current client and server.
vi...
Opens in vi any text file in the current directory.
vi macro/...
Opens in vi any text file in the global macro directory
(/usr/local/gifa/macro). Macros in this directory are available to all the Gifa
users. Equivalent to the vim macro.
vi ~/macro/...
Opens in vi any text file in the personal macro directory
($HOME/macro). Macros in this directory are available to you, wherever the
directory you are in. Equivalent to the vip macro.
OPERATION PRINCIPLE
Processing NMR data-set, is just issuing a series of correct commands along
with correct parameters. You will find starting from here a list of commands,
by topics. A list of commands by alphabetic order can be found on an other part
of the manual.
Different kind of commands
There are two kinds of commands in Gifa : regular commands which actually
execute some action (such as FT for Fourier transform), the others are commands
which may execute an action but will also change the value of internal
parameters of the program (such as LB for line broadening). Such commands will
be called contexts.
Entering spectral parameter
Most commands and macro promt for parameters. There are general rules for
parameter entering which I will try to descibe here.
General set-up
The standard way of working on an NMR spectra with Gifa is to hold everything
in memory. This is in contrast with most NMR programs that process data on disk
files, and permitted by a clever memory management.
BASIC PROCESSING
Loading and Saving data-sets : READ, WRITE, READx, WRITEx
You can load an NMR data-set by typing the READ command. It will load the
data-set in the working memory.
Apodisation : EM, LB, GM, GB, SIN, SQSIN, TM
A comprehensive set of apodisation function is available :
Zero-filling : CHSIZE
Truncation and zero-filling can be applied by the CHSIZE command which changes
the size of the data-set. You can then truncate a FID or append zeros and the
end. CHSIZE can be used on any data-set, not only on FIDs, but the EXTRACT is
preferable for spectra. The zero filling capabilities in the Graphic User
interface uses the CHSIZE command associated to the power2() function.
Fourier transform : REVF, FT, IFT, RFT, IRFT, FTBIS, IFTBIS, INVF, REVERSE
These commands are for the basic Fourier transform step.
Data-type : ITYPE
1D Data-set are either real or complex. On multidimensional data-set (2D or
3D), each direction may be either real or complex. This the so-called
hypercomplex data-type.
Phasing : PHASE, PH, HPHASE, apsl (automatic phasing)
Phasing of the data can be applied with PHASE a b. The 2 parameters a and b,
are respectively the 0th order (frequency independent) and 1st order (frequency
linearly dependent) phase corrections. In 2D and 3D, axis on which phase
correction should be applied is also entered. The pivot for the first order
correction is always the centre point of the spectrum. PH is an interactive
mode for phasing 1D data-sets. you can choose and move around your pivot (the
double line) by clicking on the spectrum with the second button of the mouse,
and you can add phase corrections using the Dialogue box. You can zoom in and
out, selecting the zoom box with the first button. Clicking "OK" will finish
the PH mode, leave the spectrum phased and load the PHASE default values with
the one you just chose, clicking "CANCEL" will exit the PH mode as if nothing
had happen. HPHASE permits to phase real data-sets, with the same syntax as
PHASE.
ADD, ADDH, ADDDATA, MULT, MULTDATA
Two data-sets can be added together with the ADD command which prompts you for
a file-name, then adds the current data-set along with the content of the file
"file-name" in standard format, the result being left in memory. It is also
possible to add a file in FT-NMR format with the ADDH command. Finally it is
also possible to add the current buffer with the data held into the DATA buffer
with the ADDDATA command (single word). In all cases, you may want to pre-scale
the current data-set with the command MULT which allows one to multiply the
current data-set by a given scalar.
Baseline correction : BCORR
BCORR is a comprehensive base-line correction module. Linear, cubic spline or
polynomial base-line correction algorithms can be selected. The 2 first modes
will use a set of points or "pivot" points to compute the base-line. A given
radius is used around each pivot point for averaging the pivot value.
EVALN, ADDBASE, NOISE, SHIFT
permits to compute the noise level and the systematic offset of the current
data-set within a given window. Computed noise level will be put into the
context NOISE, the systematic offset will be in SHIFT.
DSA
implements the Digital Shifted Acquisition method which permits reduction of
the water signal after acquisition (if the water signal is at zero-frequency).
This command will actually add the actual data with itself, after a shift and
multiplication. Values of -2 for the shift and -1.0 for the multiplication
factor will remove the water signal at zero frequency on a BRUKER data-set.
SPECTRAL PARAMETERS
SPECW, OFFSET, UNIT, FREQ
These contexts describe the spectral widths (SPECW) of the spectrum and offset
(OFFSET) of the right-most point of the current data-set, thus creating a
frequency reference for the data-set. Values are in Hz. Frequencies are used by
EM, GM, and changed by EXTRACT (but not CHSIZE, be careful.). The calib macro
permits to set the offset in an interactive manner. When no information is
available on the current data-set the OFFSET is set to 0 and the spectral
widths to 2000*Pi.
size, list
These two macros permits examination of the current state of the data-set.
size is a simple one-line text. list is a comprehensive output. (used to be
commands).
EXTRACT
permits to extract a given part of a spectrum and thus becoming the new
data-set. EXTRACT is used as ZOOM but is obviously not reversible.
DISPLAY AND GRAPHIC INTERACTION
DISP1D, DISP2D
When DISP1D is one, a 1D graphic window call "1D Spectrum" is displayed on
screen, 1D buffer will be displayed in this window . 2D will be displayed as
density colour maps in a square window call "2D window" when DISP2D is 1. In 3D
mode, planes extracted from the 3D data-sets will be displayed as 2D. Setting
these contexts to 0 actually closes the associated window.
SCALE, ITYPE, ABSMAX, SIGN, VHEIGHT, CLEAR
These contexts characterise the state of the display and the plotting.
CDISP2D, LEVEL, LOGA, CCOLOR
A second display mode is available in 2D which is a contour plot mode that is
activated with the command CDISP2D. The size of the CDISP2D window when opened
is always 15cm x 15cm. SCALE and SIGN are active in this window as they are in
the density window. The number of contour levels is defined by the LEVEL
context; LEVEL contours are displayed in the positive and in the negative part
and are equally spaced between 0 and ABSMAX/SCALE in the default mode of LOGA
equal to 1. If the context LOGA is set a value larger than 1 then each level is
at a position equal to the previous level time LOGA. For instance if with LOGA
1 levels are at :
COLOR, SCOLOR, Choice of Colours
Display are of two kinds : vector mode, used by 1D, 2D contour mode, 3D and
the SHOWxxx commands; or image used by density mode in 2D. In vector mode the
colour used is determined by the COLOR context which default value is white.
The current colour can be chosen separately for each window with the COLOR
command. Only 8 colours are available with the COLOR command, they correspond
to pure colours when the standard colour file is used. The context SCOLOR
determines the colour used by the SHOW and SHOWxxx commands (see below).
ZOOM, ZM
The ZM commands opens a control box which permits to zoom in and out the
data-set, to move around the zoom window and to rapidly set the SCALE value. To
set the zoom window click on the data-set with the left and middle button of
the mouse, and then click on the Zoom In button (or on the display, with
the left and right button of the mouse). Other controls in th ZM box are very
natural.
SHOW
permits to put some information in the graphic window. SHOW takes an argument
: CURRENT will show the current data-set as-is; FT will show the FT of it.
Other parameters are available, see below or the alphabetic list.
FREEZE
freezes the current graphic window making it inactive, and create a new one
equivalent to the previous. This is useful for comparing data.
REF, UNREF, REFMACRO
REF will force a new computing of the display, as if a display command had
been issued. This is useful for cleaning the spectrum after a point or whatever
command. UNREF realizes just the inverse, i.e. display will not be refreshed,
even if the previous commands did modify the data-set. UNREF is usefull in
macro for fine control on the display. It is active only for the commands that
preceedes it in the command line / macro ; so it is usually the last command
issued.
PLOTTING
CX, CY, ROTATE, PLOTTER, CONFIG
The 3 first commands determines the kind of plots that will be performed by
the plotting commands. The size of the plot will depend on the contexts CX and
CY (in centimetres). ROTATE exchange X and Y axes on the plotter such that the
drawings are rotated on the sheet. Two kinds of plotter can be connected to
Gifa : a HP-GL plotter (usual for pen plotter) or a Postscript (usual for laser
printers). The plotter is chosen with the PLOTTER command, you can tell for
which plotter the program is currently connected to with the CONFIG command.
PLOT, PEN, PLOT, PAGE, PLOTOFFSET, TITLE, PLOT?, FORGET
The PLOT command will plot whatever is currently on the screen. In 2D, the
plot will be strictly equivalent to what is displayed by the CDISP2D mode; in
1D, the plot will be equivalent to what is displayed, but in a CX x CY format;
in 3D the last extracted plane, as seen on the 2D display will be plotted.
PLOTAXIS, PLOTAXIS?, oe
The first command permits to draw an horizontal and/or vertical axis on the
current plot. The graduation is done according to the current values of SPECW
and OFFSET. PLOTAXIS? permits to configure the axes drawn by PLOTAXIS. GRID
permits to draw a grid of CX by CY on the current plot, it will use the axes
determined by PLOTAXIS?. Check the macros plotn and nice_plot for examples of
these functions.
STPL, STPL?
STPL will generate a stack plot either on the screen (*S), on the plotter
(*PL) or in a file. The stack-plot depends on the value of the following
contexts :
SEVERAL BUFFERS AVAILABLE
Several buffers are available in Gifa. The three main buffers (for 1D, 2D and
3D) are chosen with the DIM command. Those 3 are the only one on which generic
processing is available.
DATA ADDDATA
Is a generic buffer designed to be a "second hand". It can hold 1D, 2D or 3D
data-sets, accessed with GET and PUT, and can even be added to the current
data-set with the ADDDATA command (single word).
FILTER, WINDOW
are two 1D buffers used to manipulate the current data-set. They are mostly
designed to be used in conjunction with Maximum Entropy processing, but have
some other use however.
PUT, GET, SHOW, APPLY
are used to manipulate the different buffers.
WINDOW window used to compute the chisquare in MaxEnt processing, * * * *
also used by the polynomial mode of BCORR
FILTER {n } Generic filter function, also used for MaxEnt Deconvolution * * * *
DATA data used as a second hand by MaxEnt and Linear Prediction, * *
also used as a general purpose buffer.
LAMB the evolution of Lambda during MaxEnt iteration * *
ENT the evolution of Entropy during MaxEnt iteration * *
CHI the evolution of ChiSquare during MaxEnt iteration * *
STEP the evolution of Step during MaxEnt iteration * *
SUM the evolution of Sum of point of Image during MaxEnt * *
iteration
CONV the evolution of Convergence during MaxEnt iteration * *
RESIDUE The residue after a MaxEnt run * *
LINEFIT The result of the last line fitting * *
AMOEBA the contours used for integration during the last 2D Paris * * *
integration
FT The causal Fourier transform (FTBIS) of the current data-set *
CURRENT the current data-set (useful for comparing with SHOW) *
PLANE Fi n the nth plane of the 3D data-set, along axis Fi (F1, F2 or *
F3)
ROW n the nth row of the 2D data-set *
COL n the nth col of the 2D data-set *
HANDLING OF 2D AND 3D DATA-SETS
Introduction to 2D and 3D processing
Processing of 2D and 3D data-sets is similar to 1D data-sets in many respects.
The processing steps are mostly equivalent : reading the file, windowing,
pre-processing and zero-filling, Fourier transformation, Post-Processing
(phasing, smoothing, REAL etc...) and writing output file. This is due to the
fact that the whole data-set is merely held into central memory.
Different commands
The following command will be strictly equivalent :
Commands more specific to 2D data-sets or 3D data-set
RZOOM (2D)
When in ZOOM mode, this command will jump to the symmetrical zoom window
relative to the main diagonal of the spectrum.
COL and ROW (2D & 3D)
Permits to access directly any row (along F2) or column (along F1) of the data
set. It is put into the 1D buffer and is displayed in the 1D window. In 3D,
will extract rows and cols from the last selected plane (see PLANE).
PROJ (2D & 3D)
In 2D will compute the projection of the current data-set along one axis. Axis
along which the projection is computed has to be entered, thus PROJ F1, will
compute the projection along F1 onto F2. Two algorithms are available : mean
value or skyline. Result will be put into 1D buffer.
FLIP and FLOP (2D & 3D)
In 2D, permit to go back and forth from ITYPE=1 to ITYPE=2 data-sets by
changing the way the imaginary part is interleaved, i.e. the imaginary part is
exchanged between F1 and F2 axes. For instance the following sequence :
TRANSPOSE (2D & 3D)
transposes the 2D matrix . The sizes of the matrix must be power of two for
this command to be used. After transposition, the two dimensions are completely
permuted. In 3D, 2 dimensions will be exchanged by the TRANSPOSE process.
PLANE (3D)
Will select a plane from the 3D data-set, and put it in the 2D buffer. Plane
are select by the name of the orthogonal axis : i.e.
VERT (3D)
Will select a line orthogonal to the currently selected plane. You can choose
the coordinates by pointing on the 2D display.
DIAG (2D & 3D)
In 2D DIAG will permit to extract the main diagonal of the current data-set.
In 3D, it will extract one of the three main diagonal plane of the cube (F1=F2,
F1=F3, F2=F3). Extracting the main diagonal 1D of the 3D cube can be done by
extracting the 1D diagonal of one of the diagonal plane of the cube.
DISPLAYING AND PLOTTING 3D DATA-SETS
Plane by Plane
It is possible with Gifa to display different planes extracted from the 3D
data-set, but also to display the 3D data-set as a volume in a 3D manner.
Displaying and Plotting 3D Volumes
It is possible to display 3D pictures of 3D data set. The principle is to draw
a series of contour plots for each axes in a 3D space. This kind of display is
useful for checking the quality of a 3D data-set, and also for making nice
pictures.
DISP3D, REF3D
DISP3D will open a window devoted to 3D display. No picture is displayed when
opening because of the time a 3D display usually takes to compute. Unlike the
1D and 2D display, the 3D display must be forced with the REF3D command, and
will remain unchanged even if the data or the parameters are changed. As usual
^C permits to stop the 3D display being computed.
CX, CY, CZ, ALPHA, BETA, GAMA, AXIS3D, ZNOT, OFFSET3D, SCALE3D
This commands determine the aspect of the 3D display. The volume size is
described by the CX, CY, CZ contexts. When opening the 3D window the size is
taken to be equal to the largest of the 3 contexts. ALPHA, BETA and GAMA are
the angles which describe the view point. AXIS3D determines which planes will
be contoured. A value of F123 describes a complete contour, but simpler display
(and faster) may be chosen. Usually a F1 value is the fastest choice. ZNOT is
the focal with which the volume is displayed. A large value corresponds to a
"tele" position, a small value to a "wide". OFFSET3D determines the position of
the 0 point, values range from 0 to 1. SCALE3D is a scaling factor applied to
the final image.
DISP3D?, CHECK3D
All these parameters can be easily entered and checked with the DISP3D? macro.
An interactive, graphical mode is provided with the command CHECK3D, which
display interactively an empty cube, thus permitting to modify the main ones.
PLOT3D
This command generate on a plot file a plot equivalent to the display obtained
with the REF3D command. The plot file must then be closed with the PAGE
command, see the chapter on plotting.
WORKING ON FILE RATHER THAN IN MEMORY
Introduction to the cache system
Up to now we have only seen how the processing was performed on the data-set
currently held into the main memory. This way of working is fast and efficient
for small data-sets, but inefficient for large data-sets. So a capability has
been put into the Gifa program (since V3.0) to work on file rather than on
memory. It is thus possible to process much larger data-set, that would not fit
into the main memory.
Detailed Principle of Operation
The cache memory works on the standard file format ( accessed also through the
READ / WRITE commands). It is based on two principles : *a block structure of
2D and 3D data set files, that permits random access of data in any direction,
*a memory area that is devoted to minimising the number of disk accesses (the
cache-memory system).
Commands for using the Cache
Working with the cache system consists in creating a file in standard format
(with WRITE or with NEWFILEC), connecting to the file without actually reading
the file (with JOIN), and applying the processing either row by row (1D and 2D)
or plane by plane (3D).
JOIN, DISJOIN, LISTFILEC
The command JOIN permits to connect the program to a standard file format,
without actually loading any data into memory. The effect is to load the
contexts that describe the connected file (such as size, dimensionality, itype,
etc... see below the variable paragraph). Several files can be JOINed
independently, the contexts will always hold the parameters of the last JOINed
file. DISJOIN will disconnect the program from the currently connected file,
any modified data on the data-set will be saved onto the file. LISTFILEC output
the list of the currently JOINed file.
dataset, GETHEADER, PUTHEADER
dataset lists the value of the contexts describing the last JOINed data-set.
PUTHEADER permits to specifically modify a parameter of the currently connected
data-set, the modification is directly stored to the file(after a FLUSH or a
DISJOIN). The parameters handled by Gifa (as returned by dataset) can be
modified, but any parameter can be put into the header with this command.
GETHEADER permits to read the value of parameter in the file header. The read
value is available in the $c_header variable.
GETC, PUTC
This two commands permits to move data back and forth between the file and the
main working memory.
value of dim : 1D 2D 3D
1D 1D area 1D area 1D area
2D not applicable 2D area 2D area
3D not applicable not applicable 3D area
general
syntax is :
SHOWC
This command permits to display a currently JOINed data-set, without actually
loading it into the main memory. All the current display parameters are used
for the display, but the scale which has to be defined to the command. The
coordinates of the current zoom window used for the display is converted to
ppm, and used for the display of the JOINed data-set. This permits to display
spectra acquired in very different conditions.
FLUSH, FLUSHCACHE
The GETC, PUTC commands do not read and write data directly from the disk, but
from the cache memory. If some data are modified, the cache system takes the
burden of updating the file when needed. However, in certain cases, it might be
needed to have an updated file.
NEWFILEC
This command creates a template for a standard file format, and reserve the
room for the data. The command prompts the user for all the parameters that
will be needed to create that file. It will be then possible to fill that file
with the PUTC command.
READC, WRITEC
This two commands have been already seen in a previous paragraph. They are
strictly equivalent to the READ WRITE command. They could have been developed
as macros, using the JOIN and GETC commands for READC; and NEWFILEC and PUTC
for WRITEC.
Using macro : proc2d and proc3d
A set of macro is provided to process on file data : proc2d and proc3d in
/usr/local/gifa/macro.
proc2d :
proc2d in_file out_file axis 'list of commands'
proc3d :
proc3d in_file out_file axis 'list of commands'
UNDERSTANDING THE MEMORY SET-UP
The Gifa program holds all the data in one single large memory buffer. This
buffer is used for several purposes, and may be divided into smaller pieces.
The size of this buffer is displayed when entering the program, or with the
CONFIG command. There is no way to use a larger data-set than this memory size
but recompiling. This buffer is used for 1D as well as 2D and 3D operations. In
all cases the whole buffer is available. However, when moving back and forth
between 1D, 2D or 3D mode, or when using memory intensive commands, only
partial regions of the buffers are protected. The size of these protected
regions are given with CONFIG command. The idea is that you can eat-up all the
available memory for a single data-set if you wish, but that certain operations
will be forbidden on it.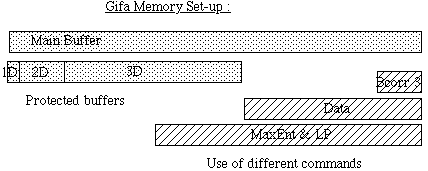
PEAK-TABLES, PEAK-PICKING LINE-FITTING AND INTEGRATION, THE PARIS MODULE
MAX, MINIMAX, PEAK
These commands permit to perform an automatic peak picking of the peak in the
current data set (1D, 2D or 3D).You have first to choose a "value-window" which
will be used by the peak-picker to select peaks in the data-set, the MINIMAX
command permits to select the upper and lower bound for this window.
Viewing : PKLIST, SHOWPEAK, SHOWPEAKS, PLOTPEAKS
These commands are the basic commands for displaying the peak table.
Storing : PKREAD, PKWRITE, PKRESET
These two commands permits to read and write peak tables on the disk. The
files created are in text format one peak per line in free format. When reading
with PKREAD, the values read can either be added to the current peak table, or
used as a new peak table. PKREAD loads all the values as stored in the file,
thus the intensities are also reloaded, the command PKRESET permits to compute
again the intensities from the peak position. PKRESET is also useful when the
data set has changed.
PKPROJ, PKSYM
These commands permit to perform mathematics on the 2D peak table. PKPROJ will
load the 1D peak table with content of the 2D peak table projected along one
dimension. PKSYM will symetrise the 2D peak table either removing or adding non
symmetrical values.
Integrating : INTEG, MSKINTEG, MODIF_AMOEBA, mskread, mskwrite
INTEG integrates all the peak found in the peak-table. This is an
implementation of the PARIS algorithm (V.Stoven et al. J.Magn.Reson. 82-
1).
The integration of a peak is performed by finding a contour spanning the
largest extension of the peak (the amoeba); the sum of the points under this
amoeba is then computed. The defined amoebae are stored as a matrix within the
program.
Line fitting : LINEFIT LINEFITD
These commands start a line fitter, based on a Levenberg-Marquardt minimiser,
convergence is determine either by the number of iteration (context ITER) or by
the size of the step (context TOLERANCE). Fitted line can be either Lorentzian
or Gaussian, as chosen by the parameter of the command LINEFIT. The content of
the peak table is used as starting values, and the result of the fit is stored
back in the peak table. The quality of the fit is estimated with a
[[chi]]2 value, which is computed as the sum of the residues :
[[chi]]2 = f(1;n) isu(i;;r(f(b(yi-Yi)2;[[sigma]]i) ))