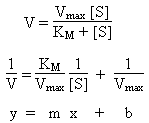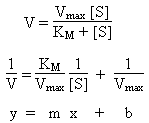Chemistry 20284 - Physiological Chemistry
Spring 1997 - Exam #2 Review
Contents
Definitions
-
Activation Energy (Ea or
DG¹)
-
Energy required to initiate a chemical reaction.
-
Active Site
-
Physical location on a protein where enzyme-catalyzed reaction occurs.
-
Allosteric Enzyme
-
Behavior of enzyme changes as a result of substrate binding.
-
Catalyst
-
A chemical compound that lowers the activation energy of a chemical reaction.
Although DG does not change, lowering
Ea increases the reaction rate.
-
Enzyme
-
Globular protein which acts as a catalyst for biological reactions.
-
Feedback Inhibition
-
Sequence of enzymatic reactions produces a substance that inhibits further
reaction.
-
Intermediate
-
Substance produced in one step of a reaction mechanism, but consumed in a
subsequent step. Not present at either beginning or end of reaction.
-
Kinetics
-
Study of rates of chemical reactions.
-
Mechanism
-
Sequence of steps required to complete a reaction.
-
Substrate
-
Reacting substance that binds to active site of an enzyme.
-
Thermodynamics
-
Study of energy changes of chemical reactions.
-
Zymogen
-
An inactive precursor that can "easily" be converted to an active form.
Rate Laws
-
General Rate Equation
-
Rate = k [A]a [B]b ...
Reaction order = a + b + ...
Order of reaction with respect to [A] = a
|
Rate = k [A]0
|
Zeroth order.
|
No dependence on [A]
|
|
Rate = k [A]1
|
First order.
|
Rate doubles if [A] doubles.
|
|
Rate = k [A]2
|
Second order.
|
Rate quadruples if [A] doubles.
|
Enzymatic Reactions
-
Lock-and-Key Mechanism
-
Enzyme assumed to be rigid. Substrate must have the exact "right" geometry
to fit in active site.
-
Induced Fit Mechanism
-
Enzyme assumed to be flexible. Geometry of enzyme can change to accommodate
substrate.
-
Competitive Inhibition
-
Inhibitor binds at active site of enzyme.
-
Non-competitive Inhibition
-
Inhibitor binds at site other than active site, which changes enzyme-substrate
affinity.
Michaelis-Menton model for Enzyme Kinetics
Plot of reaction rate (y-axis) vs. concentration of substrate
(x-axis) is hyperbolic. At low [S], first order behavior. At high [S], zeroth
order behavior. Reaction kinetics can be explained using equations
derived from the Michaelis-Menton model:
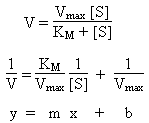
-
V
-
Reaction rate. At high concentration of substrate, approximately
Vmax. At low concentrations of substrate, follows first-order
kinetics.
-
Vmax
-
Maximum rate of reaction.
-
KM
-
Michaelis constant. Large values indicate substrate bonds weakly to enzyme.
-
[S]
-
Concentration of substrate.
-
Kinetic Analysis: Lineweaver-Burk Plot
-
Plot of V-1 (y-axis) vs. [S]-1 (x-axis)
should be linear with:
Slope = KM / Vmax
Intercept = 1 / Vmax
Nucleic Acids
-
Nucleoside
-
Consists of a base and a sugar.
-
Nucleotide
-
Consists of a base, a sugar, and a phosphate group.
-
Polynucleotide (nucleic acid)
-
Polymer of nucleotides linked by phosphodiester bonds.
-
Nucleic Acid Bases
-
Heterocyclic compounds based on either purine or pyrimidine. Contain both
H-bond donor and acceptor sites. See p 181 of text for structures.
-
Sugars
-
Nucleic acids use either ribose or deoxyribose. Both are five-carbon sugars
with a five-membered ring containing 4 C and 1 O atom.
-
Glycosidic Bond
-
N-H group of base reacts with C1-OH group of sugar to form a covalent
C-N single bond. See Figure 6.4 (p 183) in text.
-
Phosphate Group
-
PO43- reacts with C5-OH group of nucleoside
sugar to form a nucleotide. Phosphate groups can further react with the
C3-OH group of other nucleotides to form large chains linked by
phosphodiester bonds. Phosphate group maintains a negative charge.
Structure of DNA (deoxyribonucleic acid)
The primary structure of polynucleotides describes the sequence of
nucleotides present. The secondary structure describes the "local"
3-dimensional structure. For DNA, the secondary structure is a double helix,
which is driven by H-bonding between complementary base pairs (A-T and C-G).
The tertiary structure can be viewed as the "long-range" 3-dimensional
shape of the molecule. For simple (prokaryotic) cells, the tertiary structure
can be as simple as a circle. Complications include "supercoiling",
which is essentially additional twists in this ring. An example of this might
look like a "figure-8". For complex (eukaryotic) cells, the DNA tends to
wrap around positively-charged, basic proteins call histones to form
chromatin.
-
Hydrogen Bonds
-
Adenine (A) and Thymine (T) pairs form two H-bonds in DNA, while cytosine
(C) and guanine (G) pairs form three H-bonds. Increasing the relative number
of GC pairs (relative to A=T) raises the "melting point" of the DNA. See
also the "handout" on
H-bonding in DNA.
-
Double Helix
-
See p 190 of text for structure. Double helix found only in DNA, not RNA.
Negative charge of phosphate groups located on "outside" of helix. Grooves
allow substrates to "bond" to surface and access interior base pairs.
RNA (ribonucleic acid)
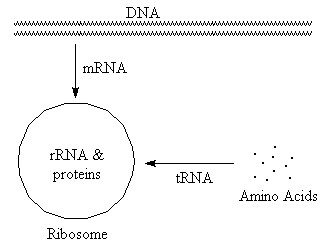
(Select text or pictures in above diagram
for additional information)
-
tRNA (transfer)
-
Carries amino acids to site of protein synthesis. Typically small, with
significant H-bonding.
-
rRNA (ribosomal)
-
Combines with proteins to form ribosomes, which are the sites of protein
synthesis. Typically large, with significant H-bonding.
-
mRNA (messenger)
-
"Translates" base sequence from DNA to ribosome. Ribosomes then use this
information to determine sequence of amino acids in protein. Variable
size, typically with very little H-bonding.
-
Structure
-
RNA is a single strand that winds back on itself to form "clover-leaf" pattern.
Contains regions where H-bonding is important and open regions where it is
not.
Restriction Enzymes
These enzymes locate specific base sequences in DNA and cleave both strands.
Sequence recognized on one strand is exact reverse of sequence on second
strand (palindrome), and cleavage occurs between same base pairs on both
strands. If breaks occur at different positions on both strands, "sticky
ends" produced. If DNA from two different sources cleaved with the same
restriction enzyme and then recombined using DNA ligase, new
(recombinant) DNA is formed. See Figures 6.22 and 6.23 of text (pp
204 & 206).
For example, a restriction enzyme might recognize the following nucleotide
sequences and cause cleavage at the positions indicated to yield the products
shown below.
|
Original DNA fragment
|
Result after cleavage
|
-----GAATTC-----
-----CTTAAG-----
|
-----G
-----CTTAA
|
AATTC-----
G-----
|
|
Note "Sticky Ends"
|
Separation and Detection Methods
-
Gel Electrophoresis
-
Method for separation of DNA or RNA fragments based on size. All fragments
move toward the positively-charged electrode, with the smaller fragments
moving faster.
-
Autoradiography
-
Incorporation of radioactive 32P makes fragment visible to X-ray
film.
-
Fluorescence
-
Fluorescent fragment (label) bonded to DNA. More sensitive method.
Determining Base Sequences in Nucleic Acids
Ribose or deoxyribose sugar portion in small percentage of nucleotides replaced
with dideoxyribose. This interrupts chain growth of growing cDNA
(complementary) strand because the C3' position no longer has
the -OH group required to react with the phosphate on the "next" nucleotide.
Fragments analyzed by gel electrophoresis. (See Figure 7.3, p 217).
Recombinant DNA
Virus or bacteria used as carrier (vector). DNA of vector and DNA
of interest cleaved by same restriction enzyme, mixed, and reformed by DNA
ligase. Recombinant DNA re-introduced in vector and grown in thin layers
to produce large number of clones. If fragments of entire DNA of organism
used, resulting clones are a DNA library. See Figures 7.11 (p 224)
and 7.13 (p 226) for examples of how these libraries are analyzed.
The polymerase chain reaction is an alternative method for creating
clones. This method is faster, and can be used to grow DNA under much less
ideal conditions.
Gene Mapping
Classically done by repeatedly comparing DNA strands broken at different
points (see Figure 7.15, p 229). More recent efforts make use of recombinant
DNA techniques (restriction enzymes, probes, and electrophoresis). Idea is
to determine how close (or far apart) select genes are from each other, and
use this information to "piece together" order of genes on DNA strand.
See also the worked out example:
Use of Restriction Enzymes for Gene Mapping.
Definitions
-
Lipids
-
An assortment of predominantly non-polar compounds that contain a limited
number of polar groups. Only marginally water-soluble.
-
Membranes
-
Formed by lipid bilayers. Polar ends on surfaces, non-polar portions
inside. Bulkier groups tend to be on outermost layer. Steroids and saturated
chains raise melting temperature, while unsaturated chains lower melting
temperature.
-
Membrane Proteins
-
Classified as integral (inside) or peripheral (outside). Have key
roles in catalysis and transport.
-
Condensation
-
Fatty acids + glycerol ¾®
triglyceride + water
-
Hydrolysis
-
Triglyceride + water ¾®
fatty acids
(If base used, reaction is referred to as saponification)
-
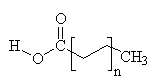
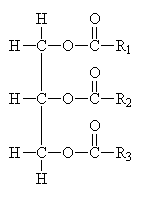
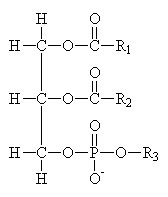
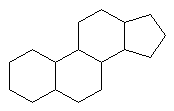
Common Classes of Lipids
|
|
|
|
|
fatty acids
|
triacylglycerides
|
phosphoacylglycerides
|
steroids
|
(Fatty acid chain lengths typically even, and run from ~12 to ~20.)
Transport of Compounds through Membranes
-
Passive
-
Movement of molecules/ions from regions of high concentration to lower
concentration.
-
Simple diffusion
-
Molecules move directly through membrane wall or a hole (channel).
-
Facilitated diffusion
-
Molecule bonds to carrier protein before transport.
-
Active
-
Movement of molecules/ions against concentration gradient. Requires
energy and a channel protein.
Section 8.7 of text on neuromuscular junctions is a good
example of membrane properties.
Includes a discussion of Voltage-gated and Ligand-gated channels.
Lipid-Soluble Vitamins
-
Vitamin A
-
cis/trans photo-induced isomerism plays a key role in vision.
-
Vitamin D
-
Important for calcium and phosphorous regulation. Derived from
cholesterol.
-
Vitamin E
-
Anti-oxidant and free-radical scavenger.
-
Vitamin K
-
Plays a key role in blood clotting.
Be able to identify isoprene units in these
compounds.
Last modified March 3, 1997
Kent State University - Stark Campus
Department of Chemistry
Dr. Clarke Earley